Figure 1.
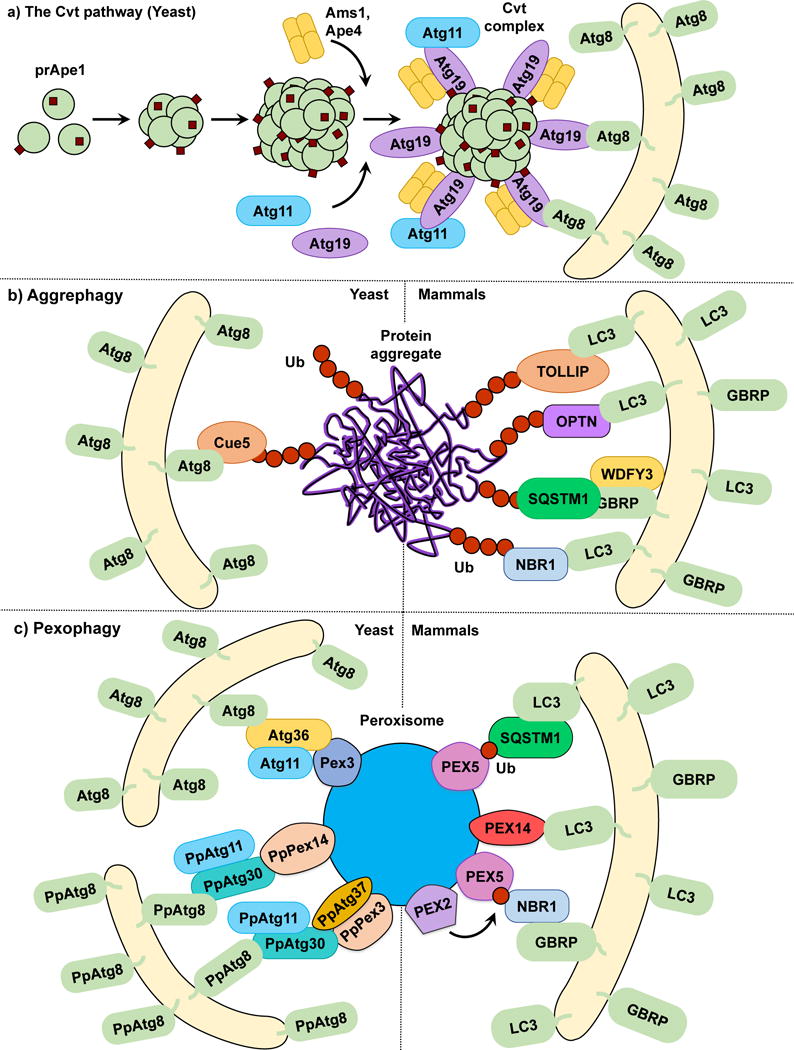
The Cvt pathway, aggrephagy and pexophagy. (a) In the yeast Cvt pathway prApe1, Ape4 and Ams1 are synthesized in the cytoplasm. prApe1 oligomerizes into dodecamers and subsequently higher order structures that are recognized by the receptor Atg19, which in turn binds the scaffold protein Atg11 forming the Cvt Complex. Ams1 and Apr4 also oligomerize and bind Atg19. Atg11 brings the Cvt Complex to the PAS where Atg19 binds Atg8‒PE, tethering the Cvt complex to the phagophore. (b) In both yeast and mammalian aggrephagy, protein aggregates are ubiquitinated and subsequently recognized by cargo receptors. In yeast, Cue5 links the ubiquitinated aggregates to Atg8‒PE. During mammalian aggrephagy, TOLLIP, SQSTM1, NBR1 and OPTN tether the ubiquitinated aggregates to the phagophore by binding LC3/GABARAP family members. WDFY3 has been described as a scaffold for SQSTM1-dependent degradation. (c) In S. cerevisiae pexophagy, Atg36 functions as a receptor linking peroxisomes to the phagophore by binding Pex3 and Atg8‒PE. In P. pastoris pexophagy, PpAtg30 acts as a receptor by linking PpPex3 and PpPex14 to PpAtg8‒PE. Atg11 functions as a scaffold for both S. cerevisiae and P. pastoris. The current model of mammalian pexophagy involves the E3-ubiquitin ligase PEX2-mediated mono-ubiquitination of PEX5, which in turn is recognized by receptors SQSTM1 and NBR1, tethering peroxisomes to the phagophore. PEX14 has also been reported to link peroxisomes to the phagophore by directly binding LC3 family members.
