Figure 5.
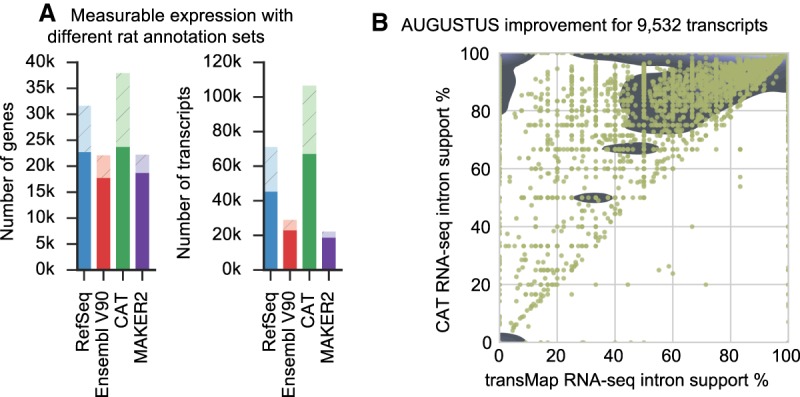
Validation of CAT annotation using rat. (A) Each transcript set was used to construct a Kallisto (Bray et al. 2016) index, and then all the input RNA-seq for annotation were quantified. Solid bars are genes or transcripts with nonzero expression (TPM>0.1) estimates, and light hatched bars are the remainder of the annotation set. CAT provides an annotation set with slightly more detectable genes than other annotation methods and far more detectable isoforms. (B) AugustusTMR provides a mechanism to clean up transcript projections and shift splice sites, fixing alignment errors as well as real evolutionary changes. Most of the 9532 AugustusTMR transcripts chosen in consensus finding show an improvement in RNA-seq support, which is one of the features used in consensus finding.
