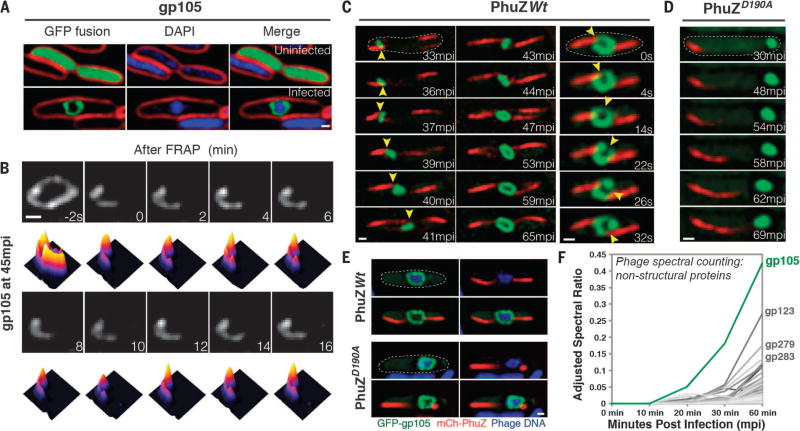
Fig. 1. gp105, an early and highly expressed phage protein, forms a shell around viral DNA during phage infection.
(A) N-terminal fusion of gp105 to GFP (GFP-gp105; green) is diffuse in uninfected P. chlororaphis cells (top) and assembles a shell that encloses the phage DNA at 45 min postinfection (mpi) with phage 201ϕ2-1 (bottom). Cell membranes were stained with FM 4–64 (red), and DNA was stained with DAPI (blue). (B) Fluorescence recovery after photobleaching (FRAP) of the GFP-gp105 shell at 45 mpi. The bleached area of the shell generated at time 0 min does not recover over the course of 16 min. Heat maps show GFP intensity corresponding to the images above. (C and D) Time-lapse imaging (measured in minutes postinfection or seconds) of GFP-gp105 in the presence of mCherry-tagged (red) (C) wild-type PhuZ and (D) mutant PhuZD190A. (C) As indicated by arrowheads, the shell moves to the midcell and rotates (in this case, clockwise) during phage infection in the presence of wild-type PhuZ. See also movies S1 and S3. (D) In the presence of PhuZD190A, the shell remains at the cell pole throughout the experiment and does not rotate. A lack of rotation is sometimes observed in the presence of wild-type PhuZ for larger gp105 shells such as that shown in (B). Dashed circles indicate the border of the cells. (E) Static images of infected host cells expressing GFP-gp105 together with either wild-type mCherry-PhuZ or mutant mCherry-PhuZD190A at 60 mpi. Phage DNA was stained blue with DAPI. (A to E) Scale bars, 0.5 µm. (F) Protein mass spectrometry analysis of phage-infected cells showing putative nonstructural phage proteins until 60 mpi. The gp105 protein (green line) is the most highly expressed phage protein. (See table S1.)