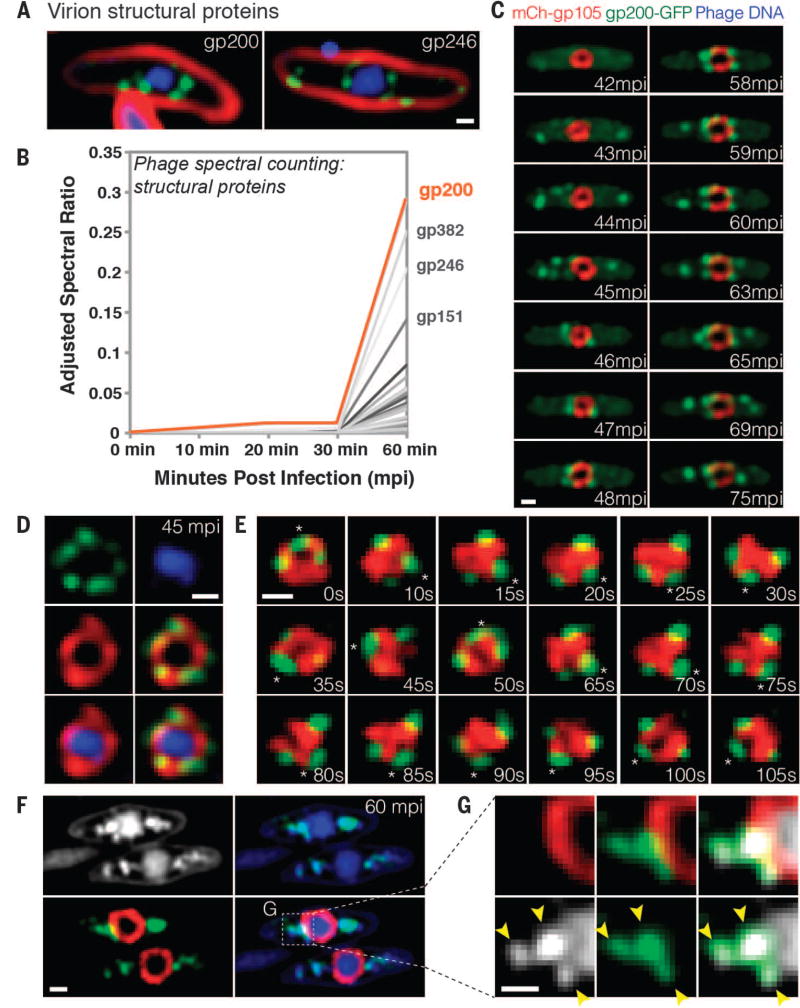
Fig. 3. Capsids migrate to the surface of the phage compartment for DNA encapsidation.
(A) Localization of the predicted phage structural proteins gp200 (major capsid protein) and gp246 (internal head protein). The proteins were fused to GFP (green), membranes were stained with FM 4–64 (red), and DNA was stained with DAPI (blue) at 60 mpi. (B) Mass spectrometry results showing spectral counting of predicted phage structural proteins in infected host cells until 60 mpi; gp200 (orange line) is the most abundant structural protein. (C) Time-lapse microscopy of mCherry-gp105 (red) and the predicted capsid protein gp200-GFP (green), showing that gp200-GFP was initially diffuse (at 42 mpi) and assembled foci on the cell periphery (at 43 to 45 mpi) that move to the gp105 shell (at 45 to 46 mpi), remain attached for 12 min, and then are released from the shell (at 59 to 75 mpi). Foci translocate to the gp105 shell within 1 to 2 min. See also movie S6. (D) Static images showing gp200-GFP (green) on the surface of the mCherry-gp105 shell (red), with DNA (blue) inside, at 45 mpi. (E) Time lapse showing that gp200-GFP foci rotate together with mCherry-gp105 throughout this 105-s experiment. The position of a single capsid (asterisk) was tracked for the duration of the time lapse. See movie S7. (F and G) Static images of infected cells at 60 mpi to show colocalization of gp200-GFP (green) and phage DNA (blue or white) outside the mCherry-gp105 shell (red). The region indicated by the dashed box in (F) is magnified in (G) to more clearly show the colocalization of DNA (white) within the gp200 foci (green). Arrowheads indicate the colocalization. Scale bars in (A) and (C) to (G), 0.5 µm.