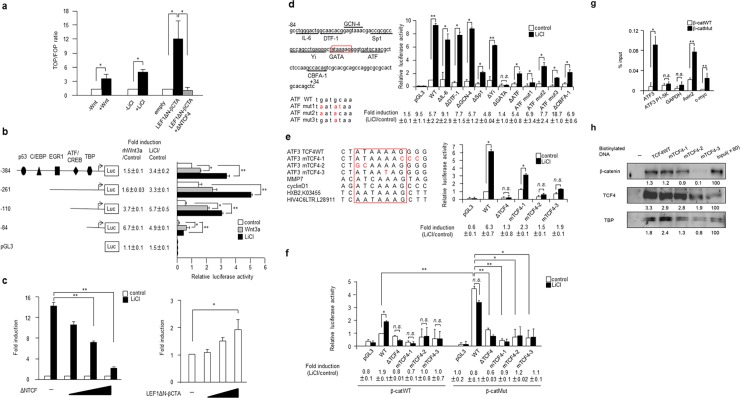
Fig 3. ATF3 is a direct target for β-catenin/TCF4 binding onto the proximal gene promoter.
(a) TOP/FOP reporter activity was assayed after the stimulation of HEK293T cells with rhWnt3a, LiCl, or co-expression of the β-catenin expression vector (LEF1ΔN-βCTA) with or without the dominant negative TCF4 plasmid (ΔNTCF4). Activity is shown as the TOP/FOP ratio relative to that of untreated or empty vector transfected cells. Data are represented as the mean ± S.E. values of three independent experiments. *, p<0.05. (b) HEK293T cells were transfected with pATF3Luc-384 or various 5′-deletion constructs of the human ATF3 reporter plasmid (left panel), and assayed for luciferase activity with or without treatment by rhWnt3a (gray columns) or LiCl (black columns). The pATF3Luc-384 plasmid contains binding elements for p53, C/EBP, EGR1, ATF/CREB, and TBP. Fold induction is the ratio of the rhWnt3a- or LiCl-stimulated activity of each reporter to that of the pATF3Luc-384 reporter plasmid without treatment. Data are represented as the mean ± S.E. values of three independent experiments. *, p<0.05 and **, p<0.01. In (c), pATF3Luc-84 was assayed for reporter activity in HEK293T cells co-transfected with increasing amounts of ΔNTCF4 (left panel) or LEF1ΔN-βCTA (right panel). Fold induction is the activity relative to that without LiCl stimulation (left panel) or to that of transfection of an empty vector. Data are represented as the mean ± S.E. values of three independent experiments. *, p<0.05 and **, p<0.01. (d) The reporter activity of various deletion or point mutants of pATF3Luc-84 (left panel) was assayed in HEK293T cells before and after LiCl stimulation, and the activity relative to that of wild type pATF3Luc-84 (WT) in the absence of LiCl is shown in the right panel. Data are represented as the mean ± S.E. values of three independent experiments. *, p<0.05 and **, p<0.01. (e) The TCF-like sequence of the ATF3 gene was aligned with those of putative genes for MMP7, cyclin D1, HXB2 and HIV4C6LTR of HIV LTR (left panel); its point mutations mTCF4-1, -2, and -3 were assayed for reporter activity in HEK293T cells before and after stimulation with LiCl (right panel). The activity relative to that of wild type pATF3Luc-84 (WT) in the absence of LiCl is shown. Data are represented as the mean ± S.E. values of three independent experiments. *, p<0.05 and **, p<0.01. In (f), various mutants of the TCF-like sequence of ATF3 gene were also assayed in Chan’s HCT116 β-catWT or β-catMut cells, as shown in (e). Data are represented as the mean ± S.E. values of three independent experiments. *, p<0.05 and **, p<0.01. (g) The β-catenin ChIP was performed in Chan’s HCT116 β-catWT (open columns) or β-catMut cells (black columns), and qPCR was performed to determine the presence of TBE (TCF4 binding motif) in the ATF3, Axin2 and c-myc genes. ATF3 P1-5K and GAPDH were the internal controls. Data are shown as the mean ± S.E. values of three independent experiments. *, p<0.05 and **, p<0.01. (h) Nuclear extracts of Chan’s HCT116 β-catMut cells were assayed using the DNAP assay as described in the Methods section, and the density of each protein was measured and its amount relative to that of the input is shown. Full-length blot images are shown in Fig e in S1 File.