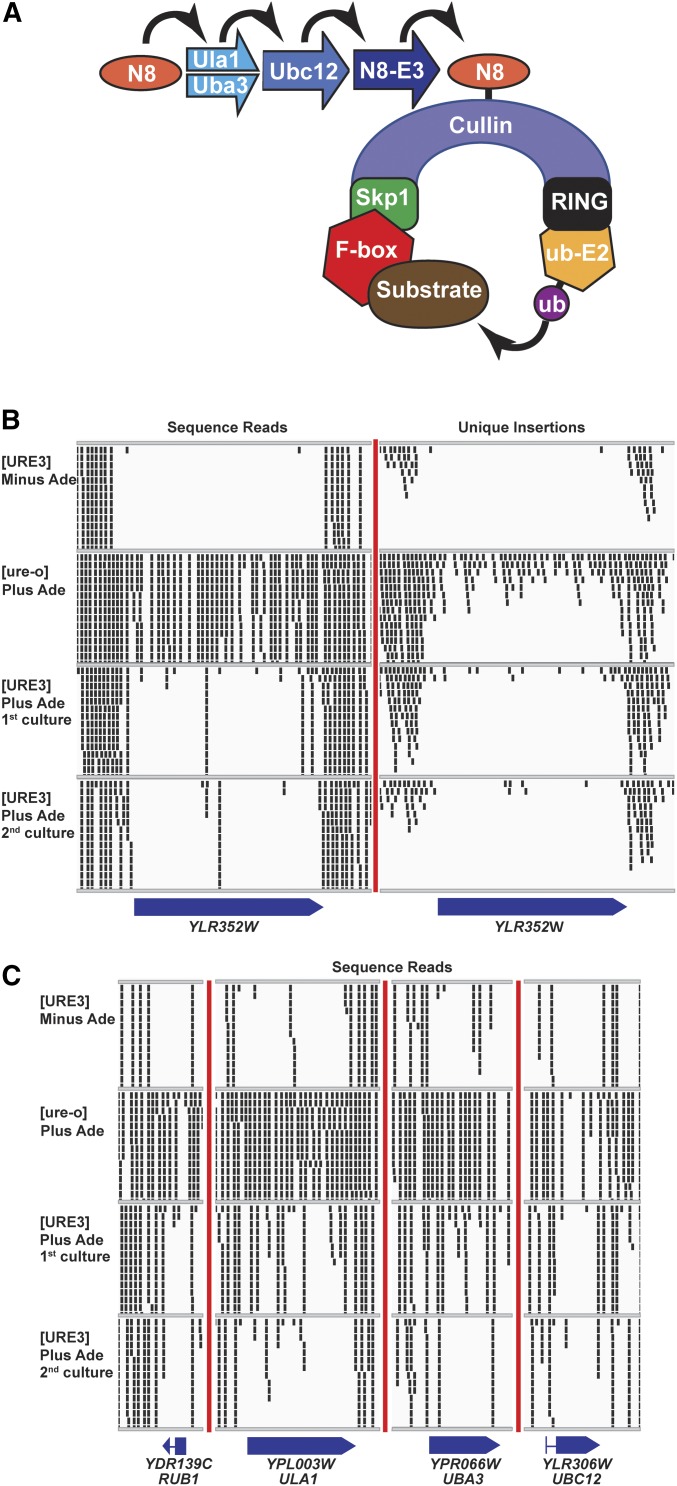
Figure 1.
(A) The SCF complex ubiquitination system. Ylr352wp/Lug1p is one of 20 yeast F-box proteins specifying substrates for ubiquitination by SCF complexes (see text). Yeast cullins include Rtt101p, Cdc53p, Cul3p, and Apc2p. The RING protein is Hrt1p. The E2-ubiquitin ligase is Cdc34p. NEDD8 (Rub1p in yeast; N8 in the figure) is a ubiquitin-like peptide whose attachment and removal from cullin is required for cycling of the SCF complex. (B) Insertions in YLR352W/LUG1 are recovered less frequently in [URE3] cells compared to [ure-o] cells. Although there were more total sequence reads for the [URE3]−Ade or [URE3]+Ade first cultures than for the [ure-o] culture, few were recovered in YLR352W/LUG1. The small rectangles represent the 50 bp to the right of the Hermes integration site. Only a maximum of 14 reads at one site, including overlapping sites, are shown. The totals in the tables include all reads/insertions. This figure is produced using IGV (http://software.broadinstitute.org/software/igv/). (C) NEDDylation genes are important for the ubiquitinylation process by SCF complexes and their mutations are also selectively underrepresented in [URE3] cultures. Total sequence reads are shown. Unique insertions are shown in Figure S5.