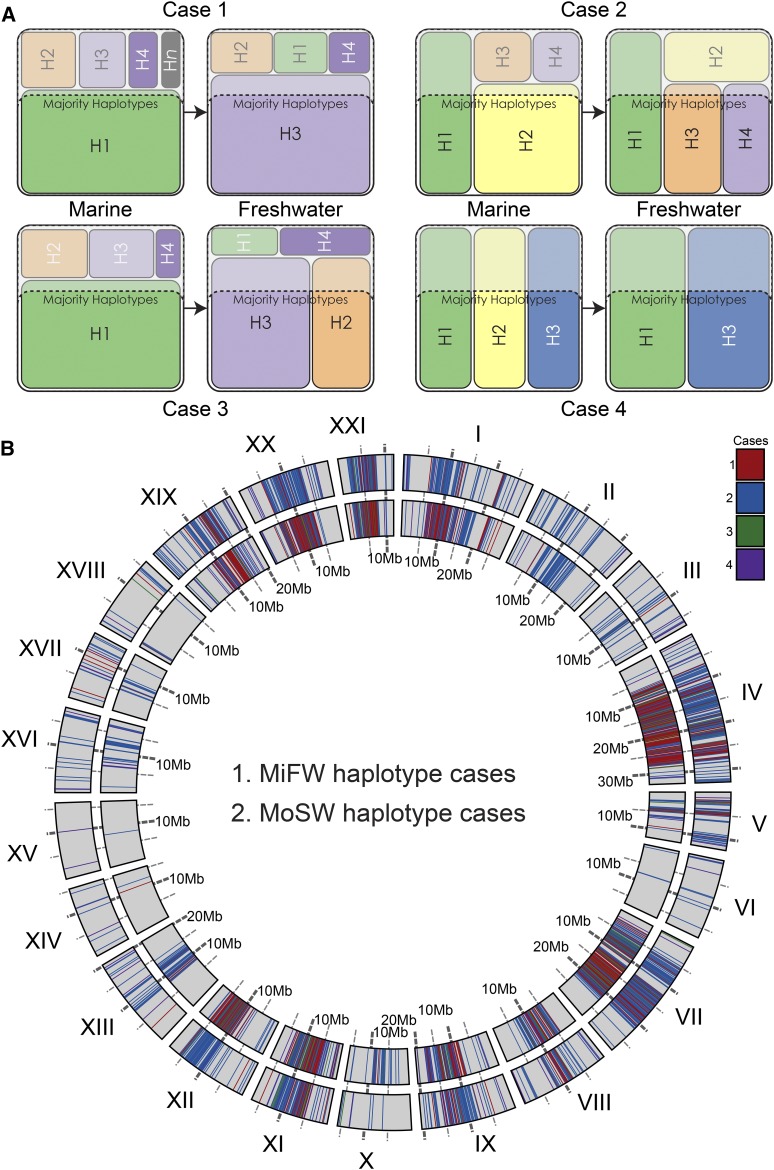
Figure 5.
Regions of parallel divergence contain loci with alternative freshwater vs. marine haplotypes. (A) To characterize the nature of loci with high FSTʹ in marine–freshwater comparisons, we first rank haplotypes at these loci from highest to lowest frequency per locus per population, define “majority” haplotypes as the highest frequency haplotypes that singly or in combination account for ≥ 60% of the population haplotypes, and then assign each locus to one of four categories: at case 1 loci, marine and freshwater populations have singular but alternative majority haplotypes; at case 2 loci, the freshwater population has one or more haplotypes that is/are majority only in fresh water, and these loci also have at least one majority haplotype that is also a majority haplotype in the sea; at case 3 loci, the freshwater population has more than one majority haplotype, none of which are majority in the sea; and at case 4 loci, the freshwater population has majority haplotypes that are a subset of the majority haplotypes in the sea. (B) Divergent loci are assigned to each of the four cases and plotted across the genome for two freshwater groups from Middleton Island (MiFW) and one from Montague Island (MoSW). Only loci with FSTʹ ≥ 0.2 are plotted, colored by case. While these cases are not strictly discreet, they broadly capture qualitatively different patterns of population genomic processes. Sampling effects could move some loci with borderline allele frequencies (i.e., those with frequencies near our parameter boundaries) between cases 1–3, but are less likely to move such loci to case 4 (for example, see Table S3).