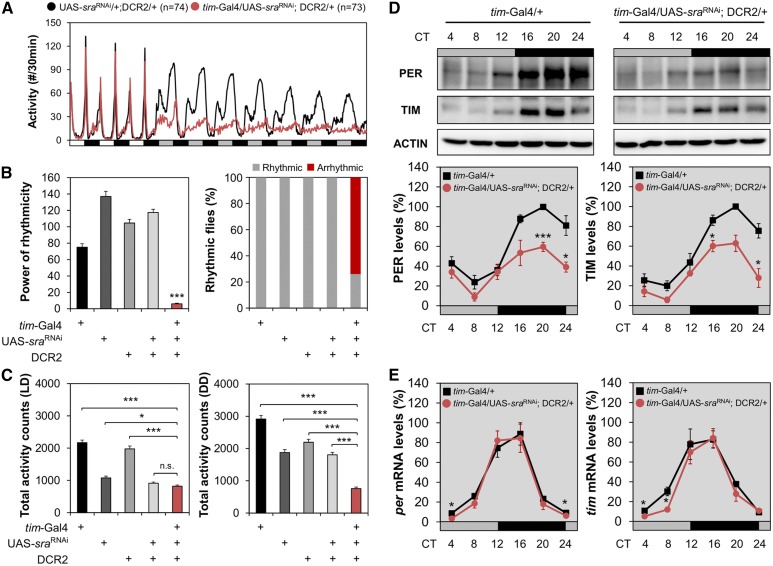
Figure 3.
sra depletion in tim-expressing circadian clock cells is sufficient to phenocopy sraKO mutation. (A) Activity profiles were averaged from individual flies in LD cycles followed by DD. (B and C) Rhythmicity in free-running locomotor behaviors [power (P) − significance (S)], percentage of flies with detectable rhythmicity (P − S > 10), and daily total activity counts were analyzed similarly as in Figure 1. * P < 0.05 and *** P < 0.001 vs. other transgenic controls, as determined by one-way ANOVA with Tukey’s post hoc test. Error bars indicate SEM (n = 31–73). (D and E) Transgenic flies were harvested at the indicated time points during the first DD cycle following the LD entrainment. Relative expression levels (%)of PER and TIM proteins (D), or those of per and tim mRNAs (E) were quantified in head extracts similarly as in Figure 2. Data represent average ± SEM (n = 3). * P < 0.05 and ** P < 0.01 vs. controls at the same time point, as determined by Student’s t-test. CT, circadian time; DD, constant dark; LD, 12 hr light/12 hr dark; n.s., not significant; UAS: upstream activating sequence.