Abstract
In vitro studies suggest that stress may generate random standing variation and that different cellular and ploidy states may evolve more rapidly under stress. Yet this idea has not been tested with pathogenic fungi growing within their host niche in vivo. Here, we analyzed the generation of both genotypic and phenotypic diversity during exposure of Candida albicans to the mouse oral cavity. Ploidy, aneuploidy, loss of heterozygosity (LOH), and recombination were determined using flow cytometry and double digest restriction site-associated DNA sequencing. Colony phenotypic changes in size and filamentous growth were evident without selection and were enriched among colonies selected for LOH of the GAL1 marker. Aneuploidy and LOH occurred on all chromosomes (Chrs), with aneuploidy more frequent for smaller Chrs and whole Chr LOH more frequent for larger Chrs. Large genome shifts in ploidy to haploidy often maintained one or more heterozygous disomic Chrs, consistent with random Chr missegregation events. Most isolates displayed several different types of genomic changes, suggesting that the oral environment rapidly generates diversity de novo. In sharp contrast, following in vitro propagation, isolates were not enriched for multiple LOH events, except in those that underwent haploidization and/or had high levels of Chr loss. The frequency of events was overall 100 times higher for C. albicans populations following in vivo passage compared with in vitro. These hyper-diverse in vivo isolates likely provide C. albicans with the ability to adapt rapidly to the diversity of stress environments it encounters inside the host.
Keywords: Candida albicans, aneuploidy, loss of heterozygosity, oropharyngeal candidiasis, hyper-variability, colony phenotype
CANDIDA albicans is a common commensal of the human gastrointestinal tract and the oral cavity in healthy individuals and is also an opportunistic pathogen, especially in immunocompromised patients (Calderone 2012). In healthy people, the fungus is prevented from causing disease by the resident microbiota and the host immune system (Lorenz et al. 2004; Richardson and Rautemaa 2009). However, immune deficiencies or a minor imbalance of the microbiota (e.g., through administration of antibiotics) can be sufficient to cause superficial infections. During the course of infection, C. albicans encounters many different host environments to which it must adapt rapidly. Furthermore, it must cope with environmental fluctuations in established niches during long-term persistence in the host (Staib et al. 2001). Determining the genetic and phenotypic changes that accompany the establishment of commensalism and the transition to pathogenicity (and hence how they can be prevented) is not known (Naglik et al. 2003; Wilson et al. 2009).
Several studies strongly suggest that C. albicans may have a very different arsenal of adaptation mechanisms when in direct contact with the host compared to laboratory conditions. For example, a novel cell phenotype (GUT) is unique to the commensal environment of the gastrointestinal tract (Pande et al. 2013) and several genes (e.g., Cph2 and Tec1) are specifically expressed under commensal conditions (Rosenbach et al. 2010). Furthermore, cells in the commensal state express genes that suggest the presence of at least two subpopulations of exponentially growing cells alongside stationary-phase cells. In addition, the expression patterns of several genes are clearly distinct during growth in vivo vs. in vitro (Nobile et al. 2006; Vandeputte et al. 2011; Fanning et al. 2012; Lohberger et al. 2014).
Recent studies in fungi found that genome instability caused by large-scale chromosomal changes; including gross chromosomal rearrangements, supernumerary chromosomes (Chrs), aneuploidy, and loss of heterozygosity (LOH); are more frequent under stress conditions in vitro and in vivo (Rustchenko et al. 1997; Selmecki et al. 2006; Coyle and Kroll 2008; Forche et al. 2009, 2011; Poláková et al. 2009; Hickman et al. 2013). Aneuploidy in particular has been shown to be one of the mechanisms that can lead to antifungal drug resistance in pathogenic fungi including Crypococcus neoformans and C. glabrata via the increase in copy number of specific genes (Selmecki et al. 2006; Poláková et al. 2009; Sionov et al. 2010). Interestingly, a recent study showed that C. albicans forms very large cells in response to acute micronutrient limitation, in particular to zinc. Cell size has been shown to be correlated with ploidy (Hickman et al. 2013) and the detection of C. albicans “Goliath” cells (Malavia et al. 2017) are consistent with an increase in ploidy as well. In C. neoformans, titan cells are large polyploid cells that can rapidly produce drug-resistant aneuploid daughters upon exposure to the drug fluconazole (Gerstein et al. 2015), supporting the idea that aneuploidy is a common adaptation mechanism of pathogenic fungi.
Our previous study of ∼80 C. albicans isolates recovered from the mouse model of hematogenously disseminated candidiasis (bloodstream infection) from mice kidneys (Forche et al. 2003, 2009) provided a first glimpse into the types of genomic changes that C. albicans undergoes at the population level. We discovered higher rates of phenotypic and Chr-level genetic variation following passage of C. albicans in vivo relative to passage in vitro. In addition, missegregation events, including whole Chr aneuploidy and LOH, were positively associated with altered colony phenotypes (CPs).
The oral cavity is one of the few host niches that is both a commensal and pathogenic niche (Vargas and Joly 2002; Patil et al. 2015). C. albicans has been found as part of the commensal microflora in up to two-thirds of the healthy population (Villar and Dongari-Bagtzoglou 2008; Pankhurst 2013). Oral and oropharyngeal candidiasis (OPC) can develop as consequence of developed immunodeficiency (e.g., HIV/AIDS), underlying diseases such as diabetes, treatment with broad-spectrum antibiotic, corticosteroids, and chemotherapy (Lyon et al. 2006; Lu et al. 2017; Sobue et al. 2018). In the oral niche, fungal–host interactions are highly dynamic due to a multitude of factors, including the presence of antimicrobial salivary peptides, the microbiota of bacterial and fungal species that coexist and compete for nutrients on epithelial cells (Demuyser et al. 2014; Jakubovics 2015) and the highly fluctuating environmental conditions (e.g., temperature and pH) (Park et al. 2009). Unexpectedly, we recently identified haploid, mating-competent C. albicans isolates for the first time, and most of these haploids were recovered after in vivo passage in an oral model of infection (Hickman et al. 2013). This extraordinary finding highlights the contribution of and the need for in vivo studies to the discovery of novel aspects of Candida biology in general and of host–pathogen interactions in particular.
To further our understanding on the acquisition of standing variation of C. albicans during infection, we performed experimental evolution of C. albicans in mice to analyze the appearance of genotypic and phenotypic diversity during passage through the mouse oral cavity for 1, 2, 3, or 5 days using an oropharyngeal model of infection (Kamai et al. 2001; Solis and Filler 2012). We found that diversity is rapidly generated after exposure to the oral host niche and that many of these changes are identified in multiple mice. The overall within-mouse diversity and multiple changes per isolate was high, independent of the duration of infection. Surprisingly, the generation of multiple genetic changes in a single isolate appears to occur with higher frequency than would be expected by random chance alone. Taken together, our results suggest that exposure to the host (and/or the transition from in vitro to in vivo growth conditions) generates highly variable isolates at a frequency two orders of magnitude higher than in vitro.
Materials and Methods
Isolate maintenance and DNA extraction
Strains used to generate parental strain YJB9318 are listed in Supplemental Material, Table S1. YJB9318 and recovered isolates were grown on YPD (2% glucose, 1% yeast extract, 1% Bacto-peptone, 20 mg/liter uridine with 1.5% agar added for plate cultures). Gal phenotypes were assessed on MIN-Gal (0.67% yeast nitrogen base without amino acids, 2% galactose, 1.5% agar; only Gal+ isolates grow) and 2-deoxygalactose medium (2DOG; 0.1% 2-deoxygalactose, 0.5% raffinose, glycerol, 1.5% agar; only Gal− isolates grow). All isolates are stored long term in 50% glycerol at −80°. DNA extractions were performed as described previously (Selmecki et al. 2005).
Construction of strain YJB9318
Plasmids and primers used in this study are listed in Table S1. YJB9318 is a derivative of strain RM1000 #2 (Table S1) in which one copy of GAL1 was replaced with URA3 (GAL1/∆gal1::URA3). First, the URA3 marker was amplified from plasmid p1374 with primers 1672 and 1673 (Table S1) and transformed into isolate YJB7617 (RM1000#2), replacing one copy of GAL1 (YJB8742) (Legrand et al. 2008). Correct disruption of GAL1 was confirmed by diagnostic PCR using primers 1674 and 1675 (Table S1). To make YJB9318 prototrophic, HIS1 was reintroduced into strain YJB8742 by transforming with plasmid p1375 (pGEM-HIS1) that was cut with restriction enzyme NruI. Diagnostic PCR with primers 728 and 565 (Table S1) confirmed correct integration of HIS1 at its native locus. To ensure that transformation did not cause any genomic changes to the parental strain, single nucleotide polymorphism (SNP) microarrays and SNP/comparative genome hybridization arrays were performed as described previously (data not shown) (Selmecki et al. 2005; Forche et al. 2009; Abbey et al. 2011).
PCR conditions for transformation and diagnostic PCR
PCRs for transformation were performed in a total volume of 50 μl with10 mM Tris–HCl (pH 8.0); 50 mM KCl; 1.5 mM MgCl2; 200 μM each of dATP, dCTP, dGTP, and dTTP; 2.5 unit rTaq polymerase (TAKARA); 4 μl of 10 μM stock solution of each primer; and 1.0 μl of template (p1374). PCRs were carried out for 34 cycles as follows: initial denaturation step for 5 min at 94°, denaturation step for 1 min at 94°, primer annealing step for 30 sec at 55°, extension step for 1 min at 72°, and a final extension step for 10 min at 72°. Each PCR product was checked by gel electrophoresis for the amplification of the desired PCR fragment. PCR products were purified using ethanol precipitation.
Diagnostic PCR was performed in a final volume of 25 μl with 10 mM Tris–HCl (pH 8.0); 50 mM KCl; 1.5 mM MgCl2; 100 μM each of dATP,dCTP, dGTP, and dTTP; 2.5 unit rTaq polymerase; 2 μl of 10 μM stock solution of each primer; and 2.5 μl genomic DNA. PCRs were carried out for 30 cycles as follows: initial denaturation for 3 min at 94°, denaturation step or 1 min at 94°, primer annealing step for 30 sec at 55°, extension step for 1 min at 72°, and a final extension step for 5 min at 72°. Five microliters of PCR product was run on a 1% agarose gel to verify that the fragment was of the appropriate size.
Model of OPC
The OPC model was essentially performed as described previously (Solis and Filler 2012). Briefly, 6-week-old male BALB/c mice (21–25 g; Taconic Farms) were immunosuppressed with cortisone acetate (225 mg/kg; Sigma Chemical, St. Louis, MO) on days −1, 1, and 3 of infection. For inoculum preparation, strain YJB9318 was grown in MIN-Gal medium to ensure that no Gal− cells arose prior infection. A total of 20 mice were infected with 1 × 106 cells of strain YJB9318 (Table 1). Of these, 17 mice survived to the scheduled dates of sacrifice. On days 1, 2, 3, and 5 postinfection (Figure 1 and Figure S1), three to five mice were killed. The tongues were extracted, weighted, and homogenized. Next, appropriate dilutions were spread onto YPD agar plates for total CFU counts and onto 2DOG agar plates to determine the number of Gal− cells. Recovered isolates were directly picked from the original YPD and 2DOG plates to 96-well plates with 50% glycerol and stored at −80° to avoid any changes to the isolates not acquired during in vivo passage.
Table 1. Frequency of CPs.
| CP | Total | Day 1 | Day 2 | Day 3 | Day 5 |
|---|---|---|---|---|---|
| Binary codea | N = 429 | N = 25 | N = 54 | N = 195 | N = 155 |
| 00 | 0.62 | 0.8 | 0.74 | 0.66 | 0.49 |
| 01 | 0.03 | 0 | 0.04 | 0.04 | 0.02 |
| 02 | 0.03 | 0.04 | 0.04 | 0.02 | 0.05 |
| 03 | 0.01 | 0 | 0.00 | 0.02 | 0.01 |
| 10 | 0.06 | 0.04 | 0.07 | 0.04 | 0.10 |
| 11 | 0.07 | 0.04 | 0.02 | 0.05 | 0.12 |
| 12 | 0.14 | 0.08 | 0.06 | 0.12 | 0.21 |
| 13 | 0.01 | 0 | 0 | 0.02 | 0.01 |
| 20 | 0.02 | 0 | 0.04 | 0.04 | 0 |
See Figure 2D for representative images.
Figure 1.
Experimental overview of in vivo and in vitro experiments.
To confirm the Gal status of recovered isolates, they were grown overnight in deep 96-well plates containing 300 μl YPD broth. Cultures were washed once with distilled water; 5 μl of each culture was spotted onto 150 × 100 mm YPD plates, MIN-Gal, and 2DOG medium; and plates were incubated for 2 days at 30° to assess growth. The frequency of LOH at the GAL1 locus was determined using the ratio of total isolates recovered (CFUs on YPD) divided by the total number of 2DOG resistant (2DOGR) isolates. The in vitro frequency of GAL1 loss in strain YJB9318 was measured as described previously (Forche et al. 2009). Of note, it is difficult to determine growth rates in vivo accurately because of bottleneck effects and because yeast cells and hyphal cells grow exponentially and linearly, respectively. Therefore, frequencies and not rates were used to compare in vitro vs. in vivo GAL1 LOH events.
Assessment of CPs and selection of isolates for genotypic analysis
Previously, we showed that isolates with missegregation events (whole Chr aneuploidy and whole Chr LOH) exhibited CPs consistent with slow growth and abnormal filamentous growth (Forche et al. 2005, 2009). To increase the ability to identify genotypic changes, we plated all isolates for CPs on YPD at 30° and scored single colonies after 3 days. CPs were determined for colony diameter (smaller or larger than parental strain; first number) and filamentous growth (degree of wrinkling compared to parental strain; second number) resulting in a binary code for each of the seven unique CPs (see Figure 2D for representative images). For further genotypic analysis, all isolates with altered CPs (6 Gal+ and 158 Gal−) and isolates with parental CP (148 Gal+ and 116 Gal−) from a total of 17 mice were chosen to yield a set of 429 isolates (Figure S1 and Table S2).
Figure 2.
Genotypic and phenotypic diversity arises early during oral infection. (A) CPs arise later in the Gal+ group (day 2, left graph) compared to the Gal− group (day 1, right graph) but are not observed in vitro. ●’s, total CFUs, ▴, CPs. (B) Ploidy changes (as measured by flow cytometry) do not arise in the Gal+ group, but do in the Gal− group with ploidy shifts observed as early as day 1 postinfection. (C) Simpson’s D is high on average and depends strongly on composition of populations within mice (Gal+:Gal− ratio of isolates). Left, overall Simpson’s D; right, Simpson’s D by days spent in mice. (D) Bubble plot shows the number of haploid, diploid, or aneuploid isolates (as measured by flow cytometry) that exhibit indicated CPs determined by growth on YPD at 30° for 3 days. CP binary codes are shown in parenthesis (see also Table S2). Bubble size reflects the number of isolates.
Determination of ploidy by flow cytometry
Ploidy of all recovered isolates was determined as described previously (Abbey et al. 2011). Briefly, each isolate was streaked out to a single colony onto YPD plates and incubated for 3 days at 30°. Single colonies were transferred to deep 96-well plates containing 0.6 ml of YPD and cultures were grown overnight (16 hr) at 300 rpm to stationary phase. Then, 50 μl were transferred to new deep 96-well plates containing 250 µl YPD broth and cultures were grown for 6 hr at 30° at 300 rpm. A total of 250 μl of culture was transferred to round-bottom 96-well plates, cells were spun down at 1000 rpm, and then resuspended in 20 µl of 50:50 buffer (50 mM Tris–HCl, pH 8.0, 50 mM EDTA, pH 8.0). To fix cells, 180 µl of 95% ethanol was added to each well. Cells were treated with 0.1 µg/ml RNase (1 hr at 37°) and 5 mg/ml Proteinase K (30 min at 37°), followed by staining with SybrGreen for 1 hr in the dark. After a final wash in 50:50 buffer, cells were resuspended in 50:50 buffer and run on a flow cytometer (FACSCALIBUR). A customized MATLAB script was used to calculate ploidy for each isolate using a diploid and a tetraploid isolate as controls (Abbey et al. 2011).
Whole-genome karyotyping using double digest restriction site-associated DNA sequencing
Double digest restriction site-associated DNA sequencing (ddRADseq) was carried out essentially as described previously (Ludlow et al. 2013) using the restriction enzymes MfeI and MboI. Sequencing was carried out on the HiSequation 2000 and NextSeq (Illumina) platforms using paired-end reads, with each read being between 39 and 50 bases in length, depending on the run. For each lane of Illumina sequencing (up to 576 isolates/lane), raw read sequences were split into isolate-specific pools based on their associated 6-bp TruSeq multiplex and 4-bp inline bar code sequences, allowing one mismatch in the i7 bar codes and no mismatches in the inline bar codes. A minimum bar code quality of Phred = 20 was applied to all bases of the inline bar code. Reads were then aligned to the C. albicans reference (SC5314 version A21-s02-m04-r01) using BWA (version 0.7.5), allowing six mismatches and quality trimming using the parameter -q 20. Because of poor base calling quality in the initial low-complexity (restriction-site) regions of reads from the NextSeq, 6 bp were trimmed from the beginning of all NextSeq reads, and this offset was recorded for identifying the position of the restriction site. The SAMtools (version 0.1.17) (Li et al. 2009) mpileup command was then used to create a pileup file for each isolate, using the -q 20 and -C 50 parameters. From the pileup file, the count of all observed bases at each covered reference position was calculated. Normalization to the euploid parent controlled for both associated restriction fragment length (which has a large impact on read depth) and GC content.
Local copy number estimation from ddRADseq:
From the aligned-read SAM file, the position of the restriction-site end of each read was determined (5′ end of forward reads, 3′ end of reverse reads), using the 6-bp NextSeq offset for NextSeq samples. Only reads with a Phred-scaled mapping alignment quality of at least 20 were considered. The occurrence of each end position was then counted, resulting in a set of marker positions for each isolate along with the number of reads aligning to each of those positions. For all isolates in a sequencing run, a matrix of observed read counts at all positions was generated. Because of the long tail of infrequently observed sites, counts of one were treated as counts of zero. This counts matrix was also used to calculate the proportion of all reads in each isolate aligning to each Chr.
Using a custom R script (File S1), first the matrix of counts was edited to remove any isolates with ≥95% positions with zero counts. Following this, marker positions were filtered to only include those occurring in >50% of remaining isolates. The edited count matrix was used to estimate relative copy number at each marker position as follows: Each isolate was normalized for depth of sequencing by dividing all observed counts by the median value of all counts greater than zero. To control for marker-to-marker variation in coverage (largely due to the size of the associated DNA fragment), normalized coverage at each marker in each isolate was divided by the median coverage (ignoring zero values) of that marker across a set of control euploid isolates. Markers called in ≤80% of euploid controls were dropped. Before plotting, globally noisy markers, with SD (of the log-normalized coverage ratios) >0.5 across all isolates, were removed and for each isolate a minimum raw count coverage of 10 or 20 was required at each position to remove low-confidence estimates for that isolate.
Ploidy was also assessed on a by-Chr basis as follows: Starting with the aligned-read proportions per Chr (see above), values for each isolate were normalized by dividing each Chr value by the median Chr value across a set of euploid control isolates. To account for variation in genome size in aneuploidy isolates, the value for each Chr was normalized (by dividing the median value for Chrs in that particular isolate). For euploid isolates, this should produce values of approximately one across all Chrs. A diploid with one trisomic Chr would have a value of ∼1.5 for one Chr and 1 for the rest. To convert these estimates to Chr copy numbers, they were multiplied by factors from 1.5:5.5 in increments of 0.1, and the factor producing the lowest root-mean-square deviation from the nearest whole number was chosen. After multiplication by this factor, Chr copy numbers were rounded to the nearest whole number. For strains known to have a median Chr copy number other than two, the set of factors was replaced by the preset median copy number ±0.5 in increments of 0.1.
For large copy number variant regions (“segmental aneuploidy”), the copy number pattern across the Chr (maximum of three segments per Chr) was identified by visualization of the marker-level copy number estimates (see above). The end points of the segments were then estimated by maximum likelihood based on the probability density of the observed normalized log coverage ratios, which were estimated from normal distributions with means equal to the log of each segmental copy number and with a common SD. The common SD was estimated for each isolate from the genome-wide log coverage ratio data using a sliding window of 11 markers.
Estimation of allele ratios:
Heterozygosity was assessed at all sites heterozygous in parental strain SC5314 (Muzzey et al. 2013), ignoring indels. For each Chr, SC5314 heterozygous sites were identified by aligning the two phased haplotypes (“A” and “B”) from Muzzey et al. (2013) using the MUMmer (version 3.22) nucmer command with the parameters -c 100 -l 10 -b 200 (Delcher et al. 2002). Alignments were filtered using the delta-filter command with the -g parameter and SNPs were then called using the show-snps command with the parameters -r -C -H -T. In each isolate, for every expected heterozygous site covered by reads, the counts of the two expected alleles were extracted from the count of all observed alleles.
The following steps were carried out by a custom R script (File S1). The binomial probability of the observed counts was calculated using models 1A:0B [homozygous A (HomA)], 3A:1B, 2A:1B, 1A:1B, 1A:2B, 1A:3B, and 0A:1B [homozygous B (HomB)]. For example, for model 3A:1B the binomial probability of the observed data would be calculated based on P(A) = 0.75, P(B) = 0.25. For the homozygous models, observed counts of the “wrong” allele were assumed to be errors with a probability of 0.01, with the expected allele having a probability of 0.99. For each site, the set of possible models was constrained based on the copy number of the Chr (or Chr segment), estimated as above. For disomic Chrs, models 1A:0B, 1A:1B, and 0A:1B were compared and the best and second-best models were identified. For trisomic Chrs, the best and second-best models were identified from models 1A:0B, 2A:1B, 1A:2B, and 0A:1B. For tetrasomes, the models compared were 1A:0B, 3A:1B, 1A:1B, 1A:3B, and 0A:1B. For any other copy number (i.e., five or above), the best of all seven models above was compared to the second-best and then final models were displayed as HomA, A > B (3A:1B or 2A:1B), 1A:1B, A < B (1A:2B or 1A:3B), HomB. After identification of the best model, for each site in each isolate, a LOD score [log10P(best model)/P(second-best model)] was calculated. For each data set, markers were removed unless they were classified as heterozygous (best model = 1A:1B with LOD greater than one) in at least one isolate. For visualization, a median sliding window of size 7 was applied to the best model values, ordered by genome position and colored by Chr/segment copy number and best model.
To obtain insight into the actual frequency of CPs and genotypic changes for the total number of CFUs counted and not just for the number of analyzed isolates, we calculated the frequencies of phenotypic and genotypic changes for Gal+ and Gal− isolates based on the total number of C. albicans CFUs from the OPC experiment [852,220 (8.52 × 105)], the number of Gal+ [851,270 (8.51 × 105; 99.89%)], and the number of Gal− cells [913 (9.5 × 102; 0.11%)]. Extrapolation showed that the actual frequency of Gal++CP was 3.9 × 10−2 (∼1/400 cells) and ∼5.8 × 10−4 for Gal−+CP (∼6/10,000 cells) (Figure S1). The frequency of genotypic changes (Chr1 changes excluded) for Gal+ cells was 13 × 10−2 and 5.2 × 10−4 for Gal− cells (Figure S1).
Diversity index
ddRADseq data were used to determine the number of unique karyotypes that were present in each mouse and the number of colonies that exhibited each karyotype. A karyotype was considered unique if there was either a unique whole or partial Chr aneuploidy or LOH event and/or there was variation in the LOH breakpoint, compared with other colonies isolated from the same mouse. Diversity was then calculated as Simpson’s index (1 − ∑pi2) (Simpson 1949), where pi is the proportional abundance of each colony type (note that if there is only one karyotype, diversity is zero).
Data availability
ddRADseq data have been deposited at National Center for Biotechnology Information Sequence Read Archive under accession numbers PRJEB24827, PRJEB24934, PRJEB24935, and PRJEB24966. Supplemental material (Figures S1–S6, Tables S1–S6, and File S1) are available at Figshare: https://doi.org/10.25386/genetics.6122360.
Results
To analyze diversification rates of C. albicans on a mucosal surface, we seeded the oral cavity of 20 corticosteroid-treated mice with 106 cells originating from YJB9318, a single, colony-purified C. albicans strain that was heterozygous for GAL1 (Gorman et al. 1992). Groups of three to five mice were sacrificed on days 1, 2, 3, and 5 and isolates were recovered from tongue homogenates (Figure 1 and Figure S1). Initiating the experiment with a Gal+/− strain enabled us to acquire evolved isolates from both YPD plates (Gal+, 541 colony isolates, no selection, unknown genomic changes) and 2DOG plates (Gal−, 360 colony isolates, minimum genomic change of LOH at the GAL1 locus) (Figure 2A and Figure S1). All recovered isolates were first screened for changes in CP and we identified seven distinct CPs, three of which were also detected among Gal+ isolates and all seven were detected among Gal− isolates (Figure 2 and Figure S1; see below). As measured by flow cytometry, the majority (72%) of isolates retained a diploid genome content (Figure 2B). Strikingly, 11 isolates had haploid or near-haploid genome (2%) content. An additional seven isolates were tetraploid or near-tetraploid while the remainder (26%) had ploidy values consistent with aneuploid diploids (Table S2).
Twenty-four hours after infection, the oral fungal burden was ∼102 CFUs per gram of tissue, suggesting that only a small proportion of the starting inoculum initiated the oral infections (Figure 2A). The number of CFUs generally increased with time of infection and the proportion with CPs increased slightly (Table 1). The proportion of Gal− CFUs increased proportionally with the total CFUs (measured by comparing the frequency of 2DOGR isolates and YPD isolates). The overall frequency of LOH at GAL1 was two orders of magnitude higher in vivo compared with in vitro (Figure S2). Ploidy changes were much more prevalent in the isolates selected for GAL1 LOH (Figure 2B) compared to isolates from YPD, and nondiploid isolates were frequently associated with reductions in both colony size and filamentous growth (Figure 2D).
Genotypic diversity by ddRADseq
ddRADseq analysis was used to analyze Chr copy number and allele frequencies from 154 isolates from YPD and 275 isolates from 2DOG. The overall diversity was calculated using Simpson’s index of diversity (1 − D) (see Materials and Methods) for each mouse. The within-mouse diversity was variable at day 1 and remained high thereafter (Figure 2C). This suggests that diversity was generated early after infection and was not highly deleterious to survival and growth within the host.
We then clone corrected the data set based upon the assumption that isolates with identical genotype and CP from the same mouse were likely to be daughter isolates resulting from a single mutational event. When the same event (genomic change) was found in different mice, we expect that event was either frequent and/or subject to strong selective pressure in the mice and was termed a “recurrent” event.
Isolates that underwent ploidy shifts based on flow cytometry were reanalyzed by ddRADseq. Interestingly, euploid shifts (the loss or gain of complete sets of Chrs) were extremely rare; only three (of 10 confirmed) haploids and none of the seven confirmed triploids or tetraploids were euploid (Figure 3, A and B). By contrast, trisomy was detected for every Chr, with higher trisomic frequencies for smaller Chrs and ChrR (Figure 4, A and B) (Table 2 and Table S3). There were seven isolates in which the majority of Chrs (more than four) were nondisomic or where Chrs were present in multiple ploidy levels (e.g., monosomy, disomy, and trisomy within the same isolate), providing indirect evidence that euploid shifts likely preceded subsequent Chr missegregation events (Figure 3C) (Forche et al. 2008; Harrison et al. 2014; Hickman et al. 2015). Importantly, aneuploidy was detected in both Gal+ and Gal− isolates (Figure S3).
Figure 3.
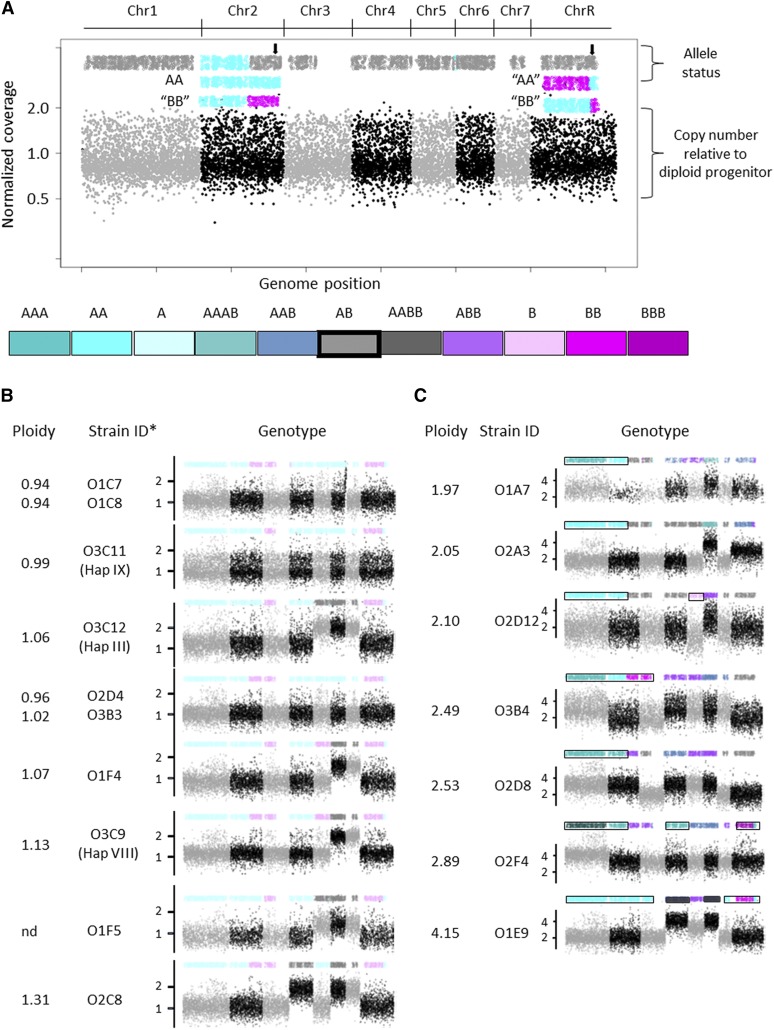
Whole-genome ploidy shifts are rare. (A) Shown is the ddRADseq whole-genome karyotype for parental strain YJB9318. Allele status is indicated on the top. Bottom half of figure provides copy number for each Chr relative to diploid parent (1 = 2 copies). Chrs are colored in light gray and black to indicate start/end of each Chr. Color coding is used throughout for indicated genotypes. Each dot on the bottom part (copy number) is a copy number estimate for a restriction fragment based on the reads aligning to one end of the restriction fragment. The dots on the top part (allele status) are maximum-likelihood estimates of allele ratios at each (sequenced) known SNP site, constrained by the Chr or segment copy number and smoothed across a number of adjacent sites (see also Materials and Methods). The colors for the allele status provide exact genotype for each Chr. Note: This strain background (RM1000 #2) has a preexisting Chr2L allele A homozygosis and a crossover on ChrR occurred during generation of the parental strain that was unmasked in isolates that became homozygous. In the case of whole Chr LOH, the genotype at the centromere was called (see black arrows). Gaps in allele coverage on Chrs3, 7, and R are due to lack of heterozygosity in the reference strain SC5314 used for analysis (Forche et al. 2004; van het Hoog et al. 2007; Butler et al. 2009). (B) Haploids and near haploids exhibit different genotypes. *, strain names in parenthesis are from Hickman et al. 2013; x-axis, Chrs are ordered Chrs1–7 and ChrR; y-axis, Chr copy number. (C) Isolates with more than two ploidies per genome suggestive of ploidy shifts in progress.
Figure 4.
Overview of missegregation events across Chrs. (A) Whole Chr aneuploidies include trisomies and tetrasomies. The number of disomic Chrs from haploids and near haploids is shown in parentheses. Y-axis: normalized copy number relative to diploid parent. Aneuploid Chrs are boxed in. Note: Images show single whole Chr aneuploidies for clarity; most isolates carry more than one whole Chr aneuploidy. (B) Single and double aneuploidies are detected both for Gal+ and Gal− isolates. Shown is number of isolates with one aneuploidy and two aneuploidies that were acquired in vivo. Chrs are shown from Chr1, Chrs2–7, and ChrR. (C) Whole Chr LOH more frequently occurs on larger Chrs1–3, and ChrR in Gal− isolates. Combinations of whole Chr LOH were not observed in Gal+ isolates.
Table 2. Summary of missegregation events by Chr.
| Chr | Whole Chr aneuploidy | Whole Chr LOH | |||||
|---|---|---|---|---|---|---|---|
| Total | 1N | 3N | 4N | Total | AA | BB | |
| Chr1 | 18 | 10 | 7 | 1 | 81 | 81 | 0 |
| Chr2 | 16 | 10 | 6 | 0 | 28 | 18 | 10 |
| Chr3 | 11 | 10 | 1 | 0 | 41 | 11 | 30 |
| Chr4 | 24 | 9 | 12 | 3 | 8 | 8 | 0 |
| Chr5 | 34 | 10 | 24 | 0 | 6 | 5 | 1 |
| Chr6 | 83 | 5 | 72 | 6 | 4 | 4 | 0 |
| Chr7 | 19 | 5 | 12 | 2 | 5 | 5 | 0 |
| ChrR | 23 | 10 | 13 | 0 | 22 | 15 | 7 |
Number (events) for each genotype are shown separately for whole Chr aneuploidy (1N, monosomic; 3N, trisomic; 4N, tetrasomic) and whole Chr LOH (AA, homozygous allele A; BB, homozygous allele B).
Haploids were detected using flow cytometry optimized after detection of an initial haploid isolate from in vitro studies (Hickman et al. 2013). The detection of multiple haploid isolates (10/950 2DOGR isolates recovered initially from the mouse oral cavity) was unexpected and exciting. Of note, only three haploids were perfectly euploid, with seven being near haploid. All the haploids tested were relatively unstable and readily converted to the autodiploid state (Hickman et al. 2015; data not shown), suggesting additional haploids may have been present in vivo. We identified nine distinct haploid or near-haploid genotypes that were recovered from six different mice: three single haploids from three different mice (Figure 3B), two unique haploids from one mouse (D3M2), three distinct haploids from one host (D5M5), and two identical haploids (D3M1; different CPs but treated as likely clones because this was the only case where we identified the same genotype for two haploids or near-haploid strains within the same mouse) (Table S2). Interestingly, only three genotypes were identical between Hickman et al. (2013) and this study, which suggests that the original isolates were a mixed population (supported by mixed flow cytometry profiles; data not shown) although the instability of haploids may have contributed as well.
Whole Chr LOH was detected for all Chrs, with higher frequencies seen for the larger Chrs (ChrR and Chrs1–3) (Figure 4C and Figure S4) (Table 2 and Table S3). The frequency of missegregation events is not entirely a function of Chr size, however, as the frequency of events on Chr3 (1.8 Mb) was higher than either Chr2 (2.1 Mb) or ChrR (2.3 Mb), which are 2.1 and 2.3 Mb, respectively. Of note, whole Chr LOH of Chrs2, 4, 6, and 7 was biased toward allele A, consistent with the failure to detect homozygous allele B in haploids or other isolates (Forche et al. 2008; Hickman et al. 2013, 2015; Ford et al. 2015; Hirakawa et al. 2015, 2017). This suggests that the B alleles of these Chrs harbor lethal recessive alleles (Feri et al. 2016) and therefore cannot be entirely lost. We did identify isolates with segmental LOH toward allele B for Chrs2, 4, and 6 (Figure 5A).
Figure 5.
Recombination and missegregation events. (A) LOH breakpoints for Chrs 2–7, and ChrR. (B) Location of LOH breakpoints along Chr1. Top horizontal black lines represent the two homologs; black oval represents centromeres; open and closed black arrows show location of the major repeat sequence; red arrows point to preexisting cross overs Chr sizes are shown to the right of each Chr. The number of isolates for each genotype are indicated at the left. For Chr1 the numbers are in shades of yellow/brown, with higher numbers shaded darker; cyan, homozygous AA; magenta, homozygous BB; gray, heterozygous AB. Breakpoints were mapped in 25-kb bins. Exact start/end coordinates of break regions can be found in Table S4. Positions 1.6–2.8 Mb on Chr1 (indicated with two solid vertical black lines) are not shown due to lack of any LOH events across this region. Maps are to scale. (C) Crossover-associated events most often lead to GAL1 loss in vivo. GC, gene conversion; missegr., missegregation; Recomb., recombination; XO, crossover.
GAL1 LOH event characterization
The isolate collection illuminates the diversity of molecular mechanisms that can yield a Gal− phenotype due to loss of the functional allele of GAL1 from the B haplotype of Chr1 (Figure 5B). We classified 264 GAL1 LOH events as either due to missegregation (involving loss of the entire B Chr) or recombination (involving LOH across the subsection of Chr1L encompassing GAL1). Recombination was more frequent than missegregation (67 vs. 33% of the total events, respectively), consistent with aneuploidy of large Chrs as rare or deleterious (Figure 5C) (Table S3). Among the Chr1 missegregation events, only the 10 A-haplotype monosomies can be explained by a single-step process: nondisjunction of the B copy of Chr1 during mitosis, leading to progeny with a single copy of the A homolog. The remaining aneuploids underwent at least two molecular events, either an increase in the copy number of the A version of Chr1 by missegregation, followed by loss of the B homolog, or vice versa. Missegregation events were much more frequent than recombination on all other Chrs, with whole Chr aneuploidy more frequent on smaller Chrs (Chrs4–7) and whole Chr LOH more prevalent on larger Chrs (Chrs1–3); on ChrR, whole Chr LOH and whole Chr aneuploidy were equally represented (Figure 5C).
Recombination events were categorized as: (1) LOH covering a Chr arm from the recombination initiation site through the telomere [likely break-induced replication (BIR)]; (2) shorter-range LOH resulting from two crossover events that do not reach the telomere (double crossovers or gene conversions); and (3) segmental copy number variations which includes truncations, amplifications, and deletions. Recombination events on Chr1L were dominated by BIR (94%), with gene-conversion events implicated in the remainder (6%; Figure 5B and Figure 6). Rare recombination events also were detected on all other Chrs in a few Gal+ as well as in Gal− isolates, and BIR was far more frequent than gene conversion (Figure 5A and Figure 6).
Figure 6.
Complex changes on individual Chrs include multiple recombination events on single Chrs (mostly Chr1), segmental deletions, truncations, and amplifications. For legend, please see Figure 5. SGM, segmental; Amp., amplification; Tr., truncation; associated black arrows point towards amplified or truncated region. For Chr1, vertical black arrows border internal segmental truncation.
The sites of recombination across Chr1L appeared relatively randomly distributed between GAL1 and CEN1 (Figure 5B). Four potential hot-spot regions were identified by binning Chr1L breakpoints every 50 kb along Chr1L (Figure S5): two to the right of the GAL1 (at ∼450 kb) locus (451–500 and 501–550 kb), one between 701 and 750 kb, and the last one between 851 and 900 kb (Figure S5 and Table S4). Most of these break regions were not near any genome elements known to promote double-strand breaks (DSBs) such as transposable elements.
In addition to the missegregation and recombination patterns described above, a small number of complex rearrangements were also detected. These included Chr truncations and recombination events that involved segmental aneuploidies and could only arise through multiple sequential events on a single Chr (Figure 6). Complex events were only seen following 2DOG selection and were most frequent on Chr1 (Figure 6) (see Table S4 for break coordinates). In addition, several isolates had multiple crossover events on Chr1, some of them involving both Chr arms with LOH to AA and BB alleles at different positions (Figure 6). Taken together, these complex genotypes suggest that DSBs were repaired via multiple, distinct mechanisms (see Discussion).
Recurrent events
The finding that the same genomic changes appeared in multiple mice supports the idea that these are general responses of the genome to conditions encountered in the oral cavity during early stages of infection. Many missegregation events occurred in multiple mice (Figure 7, Figure S6, and Table S5). Homozygosis of Chr1 to the AA genotype was seen in every mouse following 2DOG selection, presumably because this is an efficient mechanism for GAL1 LOH (Figure 4). Other recurrent whole Chr events, such as trisomy of Chr6—which appeared in the Gal+ isolates as well (Figure S6)—was unexpected. Uniparental disomy of Chr3 and Chr5 trisomy was also prevalent. Whether these different missegregation events are advantageous during early infection, or during the transition into and out of the host, remains to be determined.
Figure 7.
Recurrent missegregation events are frequent. Calculations were done for mice with C. albicans population size ≥12 (9 of 17 mice); bubble sizes reflect the percentage of mice where the specific missegregation event (indicated on x-axis) was found. For example, whole Chr1 LOH allele AA and whole Chr6 trisomy were found in all nine mice (100%). Y-axis, Chrs 1–7, and ChrR; x-axis, missegregation genotypes.
Hyper-variability in evolved isolates
Multiple combinatorial (i.e., recombination plus missegregation) events were most frequent in Gal− isolates that also exhibit CPs (Figure 8A). Not surprisingly, multiple missegregation events were found together much more frequently than multiple recombination events—likely because aneuploidies and LOH arise during concerted Chr loss from tetraploid intermediates (Forche et al. 2008; Hickman et al. 2015). Recombination events were less prevalent in general and correspondingly the frequency of multiple recombination events was also much smaller.
Figure 8.
Multiple changes (more than five) per isolate are significantly more frequent than what would be expected by random chance alone in vivo but not in vitro. (A) Multiple combinatorial (recombination + missegregation) events are most frequent in Gal− with CPs. Percentage of multiple event types for Gal+ isolates (top left), Gal+ plus CP (top right), Gal− (bottom left), and Gal− plus CP (bottom right). Y-axis, number of recombination events/isolate; x-axis, number of missegregation events per isolate. Bubble size represents the number of isolates with indicated combinations, e.g., number of isolates that have one recombination and one missegregation event. Expected vs. observed frequencies of changes (B) in vivo and (C) in vitro. Significance is indicated by * P < 0.05; ** P < 0.01.
We previously found that tetraploid isolates that underwent Chr loss yielded progeny with evidence of mitotic recombination that tended to involve multiple events on different Chrs (Forche et al. 2008). Combined with our observations of multiple and/or complex changes per isolate (Figure 5, A and B, and Figure 6), this suggests that once an isolate has undergone one mutational change it has an increased likelihood of additional changes. To quantitatively explore this idea, we calculated the frequencies of multiple vs. single events detected in the in vivo and in vitro samples (data not shown), as well as the frequency expected if each event arose randomly. The frequency of events was significantly greater than random for five or more changes per isolate for any genome change in vivo but not in vitro (only seven or more changes was significant) (Figure 8, B and C, and Table S4), indicating that in the isolates studied, highly diverse isolates are overrepresented. This implies that rare individuals undergo high levels of recombination that involve multiple Chrs.
Discussion
To understand the evolutionary forces responsible for genomic rearrangements leading to fitter genotypes, one must first identify the types of changes that reshape the genome (Chadha and Sharma 2014). Here, we provide the first population-level study of the standing variation that arises in C. albicans during OPC by analyzing several hundred isolates recovered from 17 mice at different time points during the infection. Importantly, this study design provided the perspective of time within the host. Flow cytometry and ddRADseq of 429 isolates detected many types of events due to missegregation, recombination, and multiple events of both types. Our observations of missegregation and DSB-associated changes are consistent with two recent studies of genotypic and phenotypic intraspecies variation and the evolution of drug resistance in single isolates of clinical C. albicans isolates (Ford et al. 2015; Hirakawa et al. 2015). While previous studies provide an important snapshot of ongoing changes in human infections, the lack of multiple isolates per time point makes it very difficult to recapitulate isolate genealogies throughout evolution. Of note, diversity was detectable even 1 day postinfection, suggesting that either changes arise rapidly upon the shift in growth conditions from liquid medium to the mouse and back (Jacobsen et al. 2008), or that exposure to the host environment for only 24 hr of infection is sufficient to induce genotypic changes.
The detection of multiple independent haploid or near-haploid isolates with different genotypes was surprising, suggesting that haploidization repeatedly occurs in the oral cavity. It suggests that haploidization may be advantageous during oral infection and it could lead to fitness trade-offs in subsequent generations if there is a shift in the host environment. We previously found Chr missegregation in isolates recovered after passage in a systemic model of infection and after in vitro exposure to physiologically relevant stressors (Forche et al. 2011). In vitro, the length of LOH tracts (short, long, or whole Chr) was associated with the type and severity of stress applied. Here, all three types of LOH arose at appreciable frequencies along with high levels of aneuploidy, supporting the idea that C. albicans is exposed to significant combinatorial stress in the oral cavity even though it appears to flourish in the oral cavity during OPC.
We detected a positive correlation between specific CPs and Chr missegregation. A large proportion of CPs were small in diameter and had completely smooth or less wrinkly colonies (Figure 2C), suggesting that they grow less well than the parental strain under the conditions tested and have defects in filamentous growth. This is reminiscent of the slow growth seen for aneuploid Saccharomyces cerevisiae isolates grown in laboratory media (Torres et al. 2007; Thorburn et al. 2013), which is thought to be the result of unbalanced protein stoichiometry, difficulty segregating aneuploid Chrs, or higher demands for DNA replication (Storchová et al. 2006; Torres et al. 2008, 2010; Pavelka et al. 2010; Bennett et al. 2014; Hirakawa et al. 2015).
Mutants with filamentation defects cause less damage to epithelial and endothelial cells in vitro (Phan et al. 2000; Tsuchimori et al. 2000; Bensen et al. 2002). This suggests that the isolates with reduced filamentous growth may not express hyphal-specific genes (e.g., ALS3, SAP4, and SAP6) and/or may not be recognized as readily by the host immune cells. The majority of isolates with small CPs acquired whole Chr aneuploidy, supporting the idea that these isolates may grow slowly under standard laboratory conditions, yet might have an advantage in vivo (Sem et al. 2016). Interestingly, a subset of isolates recovered after a systemic infection in mice also exhibited aneuploidy and LOH (Forche et al. 2009).
Chr6 trisomy was much more frequent than other aneuploidies, and Chr6ABB was twice as frequent as Chr6AAB. Chr6 harbors multiple members of important virulence gene families, such as secreted aspartic proteases, lipases, and adhesins (Hube et al. 2000; Naglik et al. 2004; Schaller et al. 2005; Hoyer et al. 2008; Djordjevic 2010); the NAG gene cluster important for alternative carbon utilization (Kumar et al. 2000); and RAD52, a gene important for DSB repair (Ciudad et al. 2004, 2005). Interestingly, overexpression of Rad52 increased genome instability (Takagi et al. 2008). Therefore, an extra copy of RAD52 could potentially lead to increased genome instability and amplification of specific advantageous alleles (e.g., one extra copy of allele A) could promote adaptation to specific environments such as the oral cavity. Follow-up experiments will test the effect of Chr6 trisomy on survival, persistence, and virulence of C. albicans in the oral cavity.
DSBs arise from endogenous sources including reactive oxygen species (e.g., produced by immune cells), collapsed replication forks, and from exogenous sources including chemicals that directly or indirectly damage DNA (Shrivastav et al. 2007). Using the GAL1 selection system not only allowed us to identify the major classes of genome changes and to catalog the types of LOH events that resulted in a Gal− phenotype, but it also enabled us to make hypotheses about the types of mechanisms that are involved in DSB repair. While the majority of LOH was likely the result of BIR with or without crossover, more complex LOH events also arose in a subset of isolates (see Figure 5C). The LOH signatures on Chr1 are consistent with what would be observed after short and long patch mismatch repair using different alleles as repair templates (Coïc et al. 2000; Martini et al. 2011; Bowen et al. 2013). Furthermore, more than one mismatch machinery may have been involved in repairing breaks. Strikingly, similar LOH signatures were observed during mitotic DSB repair in S. cerevisiae (Guo et al. 2017; Hum and Jinks-Robertson 2017), suggesting that these mechanisms may have been conserved through evolution.
Whether genotypic variation arises through the parasexual cycle or via mitotic defects followed by Chr missegregation accompanied by recombination events remains an outstanding question. Our previous analysis of parasexual progeny showed that the majority of them were aneuploid, that Chr missegregation predominated, that changes were observed for multiple Chrs, and that several isolates had short recombination tracts on multiple Chrs (Forche et al. 2008). A more recent study examined 32 parasexual progeny generated in vitro for a wide range of virulence-associated traits and showed that parasexual mating can generate phenotypic diversity de novo, which has important consequences for virulence and drug resistance (Hirakawa et al. 2017). Direct evidence for the parasexual cycle in vivo, however, remains elusive and the mechanism of mitotic failure followed by Chr-loss events cannot be ruled out (Harrison et al. 2014).
Importantly, here we identified a substantial level of highly variable isolates, higher than what one would expect by random chance alone. We hypothesize that hyper-variable subpopulations may be present in many natural populations, and that this diversity can enable rapid adaptation in time of stress or environmental stochasticity. Whether the observed changes are beneficial, detrimental, or neutral remains to be determined and is likely to be specific to the particulars of the environment. The link between how specific genotypic changes affects survival, persistence, and the virulence potential of C. albicans, and whether the host recognizes and responds to this variation, remains to be discovered.
Acknowledgments
We thank Carter Meyers for assistance with the animal studies. A.M.D., G.C., E.J., and A.F. were funded by National Institutes of Health grant R15 AI-090633 to A.F. S.G.F. was funded by National Institutes of Health grant R01 DE-022600 and J.B. was funded by a European Research Council Advanced Award 340087 (RAPLODAPT). A.C.G. was supported by a Canadian Institutes of Health Research Banting Postdoctoral Fellowship.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6122360.
Communicating editor: J. Stajich
Literature Cited
- Abbey D., Hickman M., Gresham D., Berman J., 2011. High-resolution SNP/CGH microarrays reveal the accumulation of loss of heterozygosity in commonly used Candida albicans strains. G3 (Bethesda) 1: 523–530. 10.1534/g3.111.000885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett R. J., Forche A., Berman J., 2014. Rapid mechanisms for generating genome diversity: whole ploidy shifts, aneuploidy, and loss of heterozygosity. Cold Spring Harb. Perspect. Med. 4: a019604 10.1101/cshperspect.a019604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensen E. S., Filler S. G., Berman J., 2002. A forkhead transcription factor is important for true hyphal as well as yeast morphogenesis in Candida albicans. Eukaryot. Cell 1: 787–798. 10.1128/EC.1.5.787-798.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen N., Smith C. E., Srivatsan A., Willcox S., Griffith J. D., et al. , 2013. Reconstitution of long and short patch mismatch repair reactions using Saccharomyces cerevisiae proteins. Proc. Natl. Acad. Sci. USA 110: 18472–18477. 10.1073/pnas.1318971110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler G., Rasmussen M. D., Lin M. F., Santos M. A. S., Sakthikumar S., et al. , 2009. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459: 657–662. 10.1038/nature08064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderone R., 2012. Candida and Candidiasis. American Society for Microbiology, Washington, DC. [Google Scholar]
- Chadha S., Sharma M., 2014. Transposable elements as stress adaptive capacitors induce genomic instability in fungal pathogen Magnaporthe oryzae. PLoS One 9: e94415 10.1371/journal.pone.0094415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciudad T., Andaluz E., Steinberg-Neifach O., Lue N. F., Gow N. A. R., et al. , 2004. Homologous recombination in Candida albicans: role of CaRad52p in DNA repair, integration of linear DNA fragments and telomere length. Mol. Microbiol. 53: 1177–1194. 10.1111/j.1365-2958.2004.04197.x [DOI] [PubMed] [Google Scholar]
- Ciudad T., Andaluz E., Steinberg-Neifach O., Lue N. F., Gow N. A., et al. , 2005. Homologous recombination in Candida albicans: role of CaRad52p in DNA repair, integration of linear DNA fragments and telomere length. Mol. Microbiol. 56: 1396 10.1111/j.1365-2958.2005.04630.x [DOI] [PubMed] [Google Scholar]
- Coïc E., Gluck L., Fabre F., 2000. Evidence for short-patch mismatch repair in Saccharomyces cerevisiae. EMBO J. 19: 3408–3417. 10.1093/emboj/19.13.3408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyle S., Kroll E., 2008. Starvation induces genomic rearrangements and starvation-resilient phenotypes in yeast. Mol. Biol. Evol. 25: 310–318. 10.1093/molbev/msm256 [DOI] [PubMed] [Google Scholar]
- Delcher A. L., Phillippy A., Carlton J., Salzberg S. L., 2002. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 30: 2478–2483. 10.1093/nar/30.11.2478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demuyser L., Jabra-Rizk M. A., Van Dijck P., 2014. Microbial cell surface proteins and secreted metabolites involved in multispecies biofilms. Pathog. Dis. 70: 219–230. 10.1111/2049-632X.12123 [DOI] [PubMed] [Google Scholar]
- Djordjevic J., 2010. Role of phospholipases in fungal fitness, pathogenicity, and drug development- lessons from Cryptococcus neoformans. Front. Microbiol. 1: 125 10.3389/fmicb.2010.00125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning S., Xu W., Solis N., Woolford C. A., Filler S. G., et al. , 2012. Divergent targets of Candida albicans biofilm regulator Bcr1 in vitro and in vivo. Eukaryot. Cell 11: 896–904. 10.1128/EC.00103-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feri A., Loll-Krippleber R., Commere P.-H., Maufrais C., Sertour N., et al. , 2016. Analysis of repair mechanisms following an induced double-strand break uncovers recessive deleterious alleles in the Candida albicans diploid genome. MBio 7: e01109-16 10.1128/mBio.01109-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forche A., May G., Beckerman J., Kauffman S., Becker J., et al. , 2003. A system for studying genetic changes in Candida albicans during infection. Fungal Genet. Biol. 39: 38–50. 10.1016/S1087-1845(02)00585-6 [DOI] [PubMed] [Google Scholar]
- Forche A., Magee P., Magee B., May G., 2004. Genome-wide single-nucleotide polymorphism map for Candida albicans. Eukaryot. Cell 3: 705–714. 10.1128/EC.3.3.705-714.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forche A., May G., Magee P. T., 2005. Demonstration of loss of heterozygosity by single-nucleotide polymorphism microarray analysis and alterations in strain morphology in Candida albicans strains during infection. Eukaryot. Cell 4: 156–165. 10.1128/EC.4.1.156-165.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forche A., Alby K., Schaefer D., Johnson A. D., Berman J., et al. , 2008. The parasexual cycle in Candida albicans provides an alternative pathway to meiosis for the formation of recombinant strains. PLoS Biol. 6: e110 10.1371/journal.pbio.0060110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forche A., Magee P. T., Selmecki A., Berman J., May G., 2009. Evolution in Candida albicans populations during a single passage through a mouse host. Genetics 182: 799–811. 10.1534/genetics.109.103325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forche A., Abbey D., Pisithkul T., Weinzierl M. A., Ringstrom T., et al. , 2011. Stress alters rates and types of loss of heterozygosity in Candida albicans. MBio 2: e00129-11 10.1128/mBio.00129-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford C. B., Funt J. M., Abbey D., Issi L., Guiducci C., et al. , 2015. The evolution of drug resistance in clinical isolates of Candida albicans. eLife 4: e00662 10.7554/eLife.00662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein A. C., Fu M. S., Mukaremera L., Li Z., Ormerod K. L., et al. , 2015. Polyploid titan cells produce haploid and aneuploid progeny to promote stress adaptation. MBio 6: e01340-15 10.1128/mBio.01340-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman J. A., Gorman J. W., Koltin Y., 1992. Direct selection of galactokinase-negative mutants of Candida albicans using 2-deoxygalactose. Curr. Genet. 21: 203–206. 10.1007/BF00336842 [DOI] [PubMed] [Google Scholar]
- Guo X., Hum Y. F., Lehner K., Jinks-Robertson S., 2017. Regulation of hetDNA length during mitotic double-strand break repair in yeast. Mol. Cell 67: 539–549.e4. 10.1016/j.molcel.2017.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison B. D., Hashemi J., Bibi M., Pulver R., Bavli D., et al. , 2014. A tetraploid intermediate precedes aneuploid formation in yeasts exposed to fluconazole. PLoS Biol. 12: e1001815 10.1371/journal.pbio.1001815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman M., Zeng G., Forche A., Hirakawa M. P., Abbey D., et al. , 2013. The ‘obligate diploid’ Candida albicans forms mating-competent haploids. Nature 494: 55–59 [corrigenda: Nature 530: 242 (2016)] 10.1038/nature11865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman M. A., Paulson C., Dudley A., Berman J., 2015. Parasexual ploidy reduction drives population heterogeneity through random and transient aneuploidy in Candida albicans. Genetics 200: 781–794. 10.1534/genetics.115.178020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa M. P., Martinez D. A., Sakthikumar S., Anderson M. Z., Berlin A., et al. , 2015. Genetic and phenotypic intra-species variation in Candida albicans. Genome Res. 25: 413–425. 10.1101/gr.174623.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa M. P., Chyou D. E., Huang D., Slan A. R., Bennett R. J., 2017. Parasex generates phenotypic diversity de novo and impacts drug resistance and virulence in Candida albicans. Genetics 207: 1195–1211. 10.1534/genetics.117.300295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyer L. L., Green C. B., Oh S. H., Zhao X., 2008. Discovering the secrets of the Candida albicans agglutinin-like sequence (ALS) gene family-a sticky pursuit. Med. Mycol. 46: 1–15. 10.1080/13693780701435317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hube B., Stehr F., Bossenz M., Mazur A., Kretschmar M., et al. , 2000. Secreted lipases of Candida albicans: cloning, characterization and expression analysis of a new gene family with at least ten members. Arch. Microbiol. 174: 362–374. 10.1007/s002030000218 [DOI] [PubMed] [Google Scholar]
- Hum Y. F., Jinks-Robertson S., 2017. Mitotic gene conversion tracts associated with repair of a defined double-strand break in Saccharomyces cerevisiae. Genetics 207: 115–128. 10.1534/genetics.117.300057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen M. D., Duncan A. D., Bain J., Johnson E. M., Naglik J. R., et al. , 2008. Mixed Candida albicans strain populations in colonized and infected mucosal tissues. FEMS Yeast Res. 8: 1334–1338. 10.1111/j.1567-1364.2008.00438.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubovics N. S., 2015. Intermicrobial interactions as a driver for community composition and stratification of oral biofilms. J. Mol. Biol. 427: 3662–3675. 10.1016/j.jmb.2015.09.022 [DOI] [PubMed] [Google Scholar]
- Kamai Y., Kubota M., Hosokawa T., Fukuoka T., Filler S. G., 2001. New model of oropharyngeal candidiasis in mice. Antimicrob. Agents Chemother. 45: 3195–3197. 10.1128/AAC.45.11.3195-3197.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M. J., Jamaluddin M. S., Natarajan K., Kaur D., Datta A., 2000. The inducible N-acetylglucosamine catabolic pathway gene cluster in Candida albicans: discrete N-acetylglucosamine-inducible factors interact at the promoter of NAG1. Proc. Natl. Acad. Sci. USA 97: 14218–14223. 10.1073/pnas.250452997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand M., Forche A., Selmecki A., Chan C., Kirkpatrick D. T., et al. , 2008. Haplotype mapping of a diploid non-meiotic organism using existing and induced aneuploidies. PLoS Genet. 4: e1 10.1371/journal.pgen.0040001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohberger A., Coste A. T., Sanglard D., 2014. Distinct roles of Candida albicans drug resistance transcription factors TAC1, MRR1, and UPC2 in virulence. Eukaryot. Cell 13: 127–142. 10.1128/EC.00245-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M. C., Bender J. A., Fink G. R., 2004. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot. Cell 3: 1076–1087. 10.1128/EC.3.5.1076-1087.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C.-W., Tao Y., Li X.-H., Dong Y., Zhou D.-D., 2017. Fungal chorioretinitis with systemic candidiasis in an infant following treatment with broad spectrum antibiotics: a case report. Exp. Ther. Med. 14: 286–288. 10.3892/etm.2017.4507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludlow C. L., Scott A. C., Cromie G. A., Jeffery E. W., Sirr A., et al. , 2013. High-throughput tetrad analysis. Nat. Methods 10: 671–675. 10.1038/nmeth.2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon J. P., da Costa S. C., Totti V. M. G., Munhoz M. F. V., de Resende M. A., 2006. Predisposing conditions for Candida spp. carriage in the oral cavity of denture wearers and individuals with natural teeth. Can. J. Microbiol. 52: 462–467. 10.1139/w05-148 [DOI] [PubMed] [Google Scholar]
- Malavia D., Lehtovirta-Morley L. E., Alamir O., Weiß E., Gow N. A. R., et al. , 2017. Zinc limitation induces a hyper-adherent goliath phenotype in Candida albicans. Front. Microbiol. 8: 2238 10.3389/fmicb.2017.02238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini E., Borde V., Legendre M., Audic S., Regnault B., et al. , 2011. Genome-wide analysis of heteroduplex DNA in mismatch repair–deficient yeast cells reveals novel properties of meiotic recombination pathways. PLoS Genet. 7: e1002305 10.1371/journal.pgen.1002305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzzey D., Schwartz K., Weissman J., Sherlock G., 2013. Assembly of a phased diploid Candida albicans genome facilitates allele-specific measurements and provides a simple model for repeat and indel structure. Genome Biol. 14: R97 10.1186/gb-2013-14-9-r97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naglik J., Albrecht A., Bader O., Hube B., 2004. Candida albicans proteinases and host/pathogen interactions. Cell. Microbiol. 6: 915–926. 10.1111/j.1462-5822.2004.00439.x [DOI] [PubMed] [Google Scholar]
- Naglik J. R., Challacombe S. J., Hube B., 2003. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol. Mol. Biol. Rev. 67: 400–428. 10.1128/MMBR.67.3.400-428.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile C. J., Andes D. R., Nett J. E., Smith F. J., Yue F., et al. , 2006. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog. 2: e63 10.1371/journal.ppat.0020063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pande K., Chen C., Noble S. M., 2013. Passage through the mammalian gut triggers a phenotypic switch that promotes Candida albicans commensalism. Nat. Genet. 45: 1088–1091. 10.1038/ng.2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankhurst C. L., 2013. Candidiasis (oropharyngeal). BMJ Clinical Evidence 2013: 1304. [PMC free article] [PubMed] [Google Scholar]
- Park H., Liu Y., Solis N., Spotkov J., Hamaker J., et al. , 2009. Transcriptional responses of Candida albicans to epithelial and endothelial cells. Eukaryot. Cell 8: 1498–1510. 10.1128/EC.00165-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil S., Rao R. S., Majumdar B., Anil S., 2015. Clinical appearance of oral Candida infection and therapeutic strategies. Front. Microbiol. 6: 1391 10.3389/fmicb.2015.01391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelka N., Rancati G., Zhu J., Bradford W. D., Saraf A., et al. , 2010. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature 468: 321–325. 10.1038/nature09529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan Q. T., Belanger P. H., Filler S. G., 2000. Role of hyphal formation in interactions of Candida albicans with endothelial cells. Infect. Immun. 68: 3485–3490. 10.1128/IAI.68.6.3485-3490.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poláková S., Blume C., Zárate J. A., Mentel M., Jørck-Ramberg D., et al. , 2009. Formation of new chromosomes as a virulence mechanism in yeast Candida glabrata. Proc. Natl. Acad. Sci. USA 106: 2688–2693. 10.1073/pnas.0809793106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson M., Rautemaa R., 2009. How the host fights against Candida infections. Front. Biosci. 14: 4363–4375. 10.2741/s24 [DOI] [PubMed] [Google Scholar]
- Rosenbach A., Dignard D., Pierce J. V., Whiteway M., Kumamoto C. A., 2010. Adaptations of Candida albicans for growth in the mammalian intestinal tract. Eukaryot. Cell 9: 1075–1086. 10.1128/EC.00034-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustchenko E. P., Howard D. H., Sherman F., 1997. Variation in assimilating functions occurs in spontaneous Candida albicans mutants having chromosomal alterations. Microbiology 143: 1765–1778. 10.1099/00221287-143-5-1765 [DOI] [PubMed] [Google Scholar]
- Schaller M., Borelli C., Korting H., Hube B., 2005. Hydrolytic enzymes as virulence factors of Candida albicans. Mycoses 48: 365–377. 10.1111/j.1439-0507.2005.01165.x [DOI] [PubMed] [Google Scholar]
- Selmecki A., Bergmann S., Berman J., 2005. Comparative genome hybridization reveals widespread aneuploidy in Candida albicans laboratory strains. Mol. Microbiol. 55: 1553–1565. 10.1111/j.1365-2958.2005.04492.x [DOI] [PubMed] [Google Scholar]
- Selmecki A., Forche A., Berman J., 2006. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 313: 367–370. 10.1126/science.1128242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sem X., Le G. T. T., Tan A. S. M., Tso G., Yurieva M., et al. , 2016. β-glucan exposure on the fungal cell wall tightly correlates with competitive fitness of Candida species in the mouse gastrointestinal tract. Front. Cell. Infect. Microbiol. 6: 186 10.3389/fcimb.2016.00186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastav M., De Haro L. P., Nickoloff J. A., 2007. Regulation of DNA double-strand break repair pathway choice. Cell Res. 18: 134–147. 10.1038/cr.2007.111 [DOI] [PubMed] [Google Scholar]
- Simpson E. H., 1949. Measurement of diversity. Nature 163: 688 10.1038/163688a0 [DOI] [Google Scholar]
- Sionov E., Lee H., Chang Y. C., Kwon-Chung K. J., 2010. Cryptococcus neoformans overcomes stress of azole drugs by formation of disomy in specific multiple chromosomes. PLoS Pathog. 6: e1000848 10.1371/journal.ppat.1000848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobue T., Bertolini M., Thompson A., Peterson D. E., Diaz P. I., et al. , 2018. Chemotherapy-induced oral mucositis and associated infections in a novel organotypic model. Mol. Oral Microbiol. 33: 212–223. 10.1111/omi.12214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solis N. V., Filler S. G., 2012. Mouse model of oropharyngeal candidiasis. Nat. Protoc. 7: 637–642. 10.1038/nprot.2012.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staib P., Wirsching S., Strauss A., Morschhauser J., 2001. Gene regulation and host adaptation mechanisms in Candida albicans. Int. J. Med. Microbiol. 291: 183–188. 10.1078/1438-4221-00114 [DOI] [PubMed] [Google Scholar]
- Storchová Z., Breneman A., Cande J., Dunn J., Burbank K., et al. , 2006. Genome-wide genetic analysis of polyploidy in yeast. Nature 443: 541–547. 10.1038/nature05178 [DOI] [PubMed] [Google Scholar]
- Takagi Y., Akada R., Kumagai H., Yamamoto K., Tamaki H., 2008. Loss of heterozygosity is induced in Candida albicans by ultraviolet irradiation. Appl. Microbiol. Biotechnol. 77: 1073–1082. 10.1007/s00253-007-1252-x [DOI] [PubMed] [Google Scholar]
- Thorburn R. R., Gonzalez C., Brar G. A., Christen S., Carlile T. M., et al. , 2013. Aneuploid yeast strains exhibit defects in cell growth and passage through START. Mol. Biol. Cell 24: 1274–1289. 10.1091/mbc.e12-07-0520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres E. M., Sokolsky T., Tucker C. M., Chan L. Y., Boselli M., et al. , 2007. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317: 916–924. 10.1126/science.1142210 [DOI] [PubMed] [Google Scholar]
- Torres E. M., Williams B. R., Amon A., 2008. Aneuploidy: cells losing their balance. Genetics 179: 737–746. 10.1534/genetics.108.090878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres E. M., Dephoure N., Panneerselvam A., Tucker C. M., Whittaker C. A., et al. , 2010. Identification of aneuploidy-tolerating mutations. Cell 143: 71–83. 10.1016/j.cell.2010.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchimori N., Sharkey L. L., Fonzi W. A., French S. W., Edwards J. E., et al. , 2000. Reduced virulence of HWP1-deficient mutants of Candida albicans and their interactions with host cells. Infect. Immun. 68: 1997–2002. 10.1128/IAI.68.4.1997-2002.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandeputte P., Ischer F., Sanglard D., Coste A., 2011. In vivo systematic analysis of Candida albicans Zn2-Cys6 transcription factors mutants for mice organ colonization. PLoS One 6: e26962 10.1371/journal.pone.0026962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van het Hoog M., Rast T. J., Martchenko M., Grindle S., Dignard D., et al. , 2007. Assembly of the Candida albicans genome into sixteen supercontigs aligned on the eight chromosomes. Genome Biol. 8: R52 10.1186/gb-2007-8-4-r52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas K. G., Joly S., 2002. Carriage frequency, intensity of carriage, and strains of oral yeast species vary in the progression to oral candidiasis in human immunodeficiency virus-positive individuals. J. Clin. Microbiol. 40: 341–350. 10.1128/JCM.40.2.341-350.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar C. C., Dongari-Bagtzoglou A., 2008. Immune defence mechanisms and immunoenhancement strategies in oropharyngeal candidiasis. Expert Rev. Mol. Med. 10: e29 10.1017/S1462399408000835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D., Thewes S., Zakikhany K., Fradin C., Albrecht A., et al. , 2009. Identifying infection-associated genes of Candida albicans in the postgenomic era. FEMS Yeast Res. 9: 688–700. 10.1111/j.1567-1364.2009.00524.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
ddRADseq data have been deposited at National Center for Biotechnology Information Sequence Read Archive under accession numbers PRJEB24827, PRJEB24934, PRJEB24935, and PRJEB24966. Supplemental material (Figures S1–S6, Tables S1–S6, and File S1) are available at Figshare: https://doi.org/10.25386/genetics.6122360.