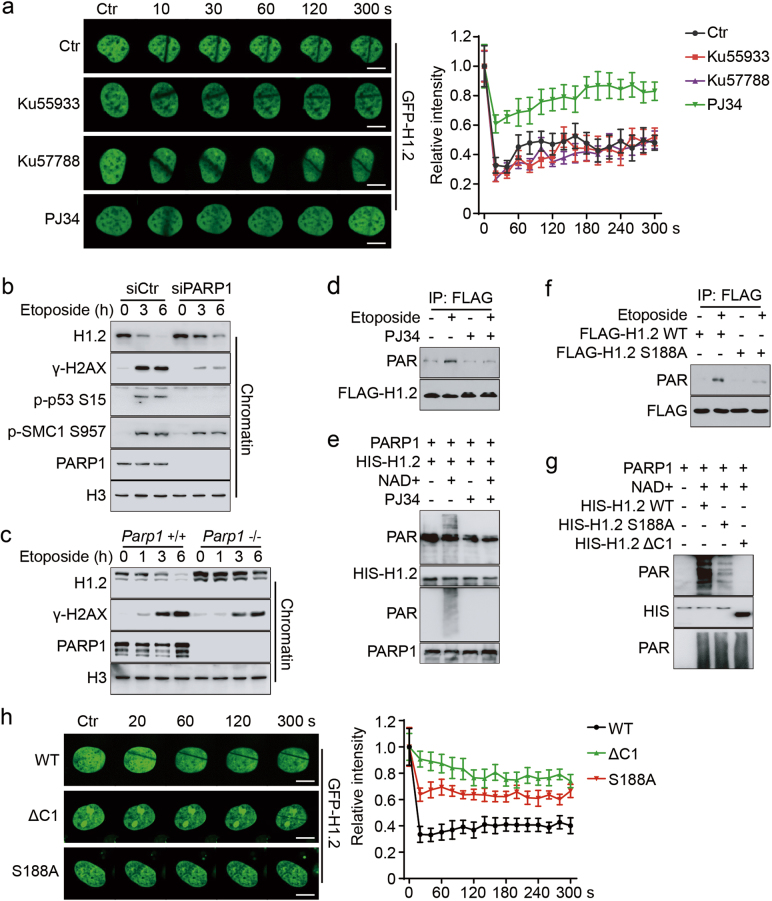
Fig. 5.
H1.2 PARylation permits its displacement from chromatin upon DNA damage. a HeLa cells were transfected with GFP-H1.2 and treated with 20 μM Ku55933 or 2 μM Ku57788 for 4 h or 5 μM PJ34 for 1 h followed by laser micro-irradiation. Images were taken every 10 s for 5 min and quantifications of the IR path signal intensity were shown and ~15 IR paths from 10 separate cells were calculated. The data represent the mean ± SD. Scale bars, 10 μm. b HeLa cells were transfected with the indicated siRNAs and treated with 40 μM etoposide for the indicated time. Chromatin was fractionated and analyzed by immunoblotting. c Parp1 wild-type (+/+) or KO (−/−) MEFs were treated with 40 μM etoposide for the indicated time and chromatin was fractionated and analyzed by immunoblotting. d HeLa cells were transfected with FLAG-H1.2 and treated with 40 μM etoposide for 15 min with or without 5 μM PJ34 for 1 h. Cell extracts were immunoprecipitated with FLAG-conjugated M2 beads. e Recombinant HIS-H1.2 was subjected to in vitro PARylation assay in the presence of NAD+ or 10 μM PJ34, as indicated. f HeLa cells were transfected with wild-type or S188A mutated FLAG-H1.2 and treated with 40 μM etoposide for 15 min with or without 5 μM PJ34 for 1 h, as indicated. Cells were extracted and immunoprecipitated with FLAG-conjugated M2 beads. g Recombinant wild-type, S188A mutated or C1-deleted (ΔC1) HIS-H1.2 were subjected to in vitro PARylation assay. h HeLa cells were transfected with wild-type, ΔC1 or S188A mutated GFP-H1.2 and subjected to laser micro-irradiation. Images were taken every 20 s for 5 min and representative images were shown. Quantifications were calculated as in a. The data represent the mean ± SD. Scale bars, 10 μm