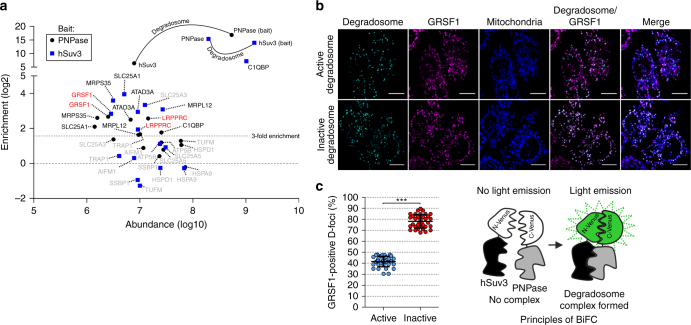
Fig. 1.
GRSF1 associates with the degradosome. a Mitochondrial proteins identified in all three purifications of hSuv3 or PNPase. Proteins that did not meet the criteria for consideration as putative degradosome-associated are labeled in gray. b Colocalization studies of the degradosome and GRSF1. The degradosome was visualized using a bimolecular fluorescence complementation (BiFC) approach, the principles of which are illustrated schematically. Catalytically active or inactive (hSuv3-G207V, PNPase-R445ER446E) components of the degradosome were transiently expressed in fusion with a split Venus protein. D-foci are not present in every cell due to transfection efficiency below 100%. GRSF1 was detected using immunofluorescence labeling. Specificity of antibodies for GRSF1 was confirmed by their pre-incubation with purified GRSF1 (Supplementary Fig. 1b). Mitochondria were stained with MitoTracker. Scale bar represents 10 µm. c Quantitative analysis of fluorescence microscopy data wherein 35 randomly selected cells were analyzed. Fraction of D-foci co-localizing with GRSF1 was calculated using the object-based colocalization approach. Individual values are shown. Horizontal lines represent mean values. Error bars represent standard deviation. A two-tailed unpaired t-test was applied (***P < 0.0001)