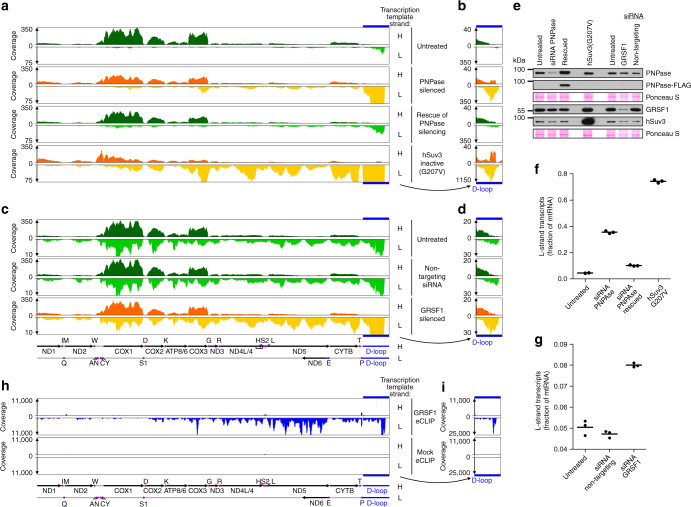
Fig. 2.
Dysfunction of the degradosome or GRSF1 upregulates RNAs originating from L-strand transcription. a–d Tracks of mapped reads of long transcript libraries. RNA from two set of experiments (a–b and c–d) performed in triplicate was analyzed. Panels b and d show the D-loop region with an adjusted scale. siRNA silencing of endogenous PNPase expression was rescued by expression of an exogenous siRNA-insensitive version of PNPase-FLAG. tRNA genes are labeled with single-letter symbols. S1 and S2 correspond to tRNA-Ser(UCN) and tRNA-Ser(AGY), respectively. e Western blot analysis of indicated proteins. Exogenous, siRNA-insensitive PNPase was detected with anti-FLAG antibodies. Membranes were stained with Ponceau S as a loading control. f, g Contribution of L-strand templated transcripts to the mitochondrial transcriptome. Individual values are shown. Horizontal lines represent mean values. h–i Distribution of GRSF1-binding sites across mitochondrial transcripts. eCLIP data from ENCODE database were re-analyzed. The D-loop region is shown with an adjusted scale (i)