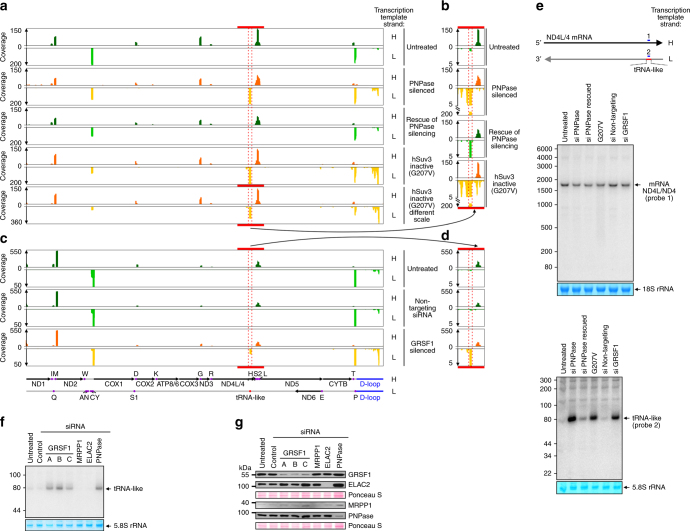
Fig. 3.
Silencing of GRSF1 or inactivation of the degradosome results in tRNA-like accumulation. a–d Tracks of mapped reads of short RNA libraries. The same RNA as that for Fig. 2a–d, f, g was analyzed. Position of newly identified tRNA-like RNA species is indicated by red dots. Panels b and d show the tRNA-like encoding region with a modified scale to visualize this RNA in control cells. S1 and S2 correspond to tRNA-Ser(UCN) and tRNA-Ser(AGY), respectively. e Confirmation of short RNA-seq results using northern blot hybridization. Strand-specific oligonucleotide probes were applied. A diagram of the probes is shown at the top. Methylene blue staining of rRNA is shown as a loading control. f Northern blot analysis of tRNA-like levels in untreated or siRNA-transfected cells. Three different siRNAs specific for GRSF1 were used. Methylene blue staining of rRNA is shown as a loading control. g Silencing of the indicated genes was confirmed by western blot analysis. Ponceau S staining of the western blot membranes is shown as a loading control. See also Supplementary Fig. 4