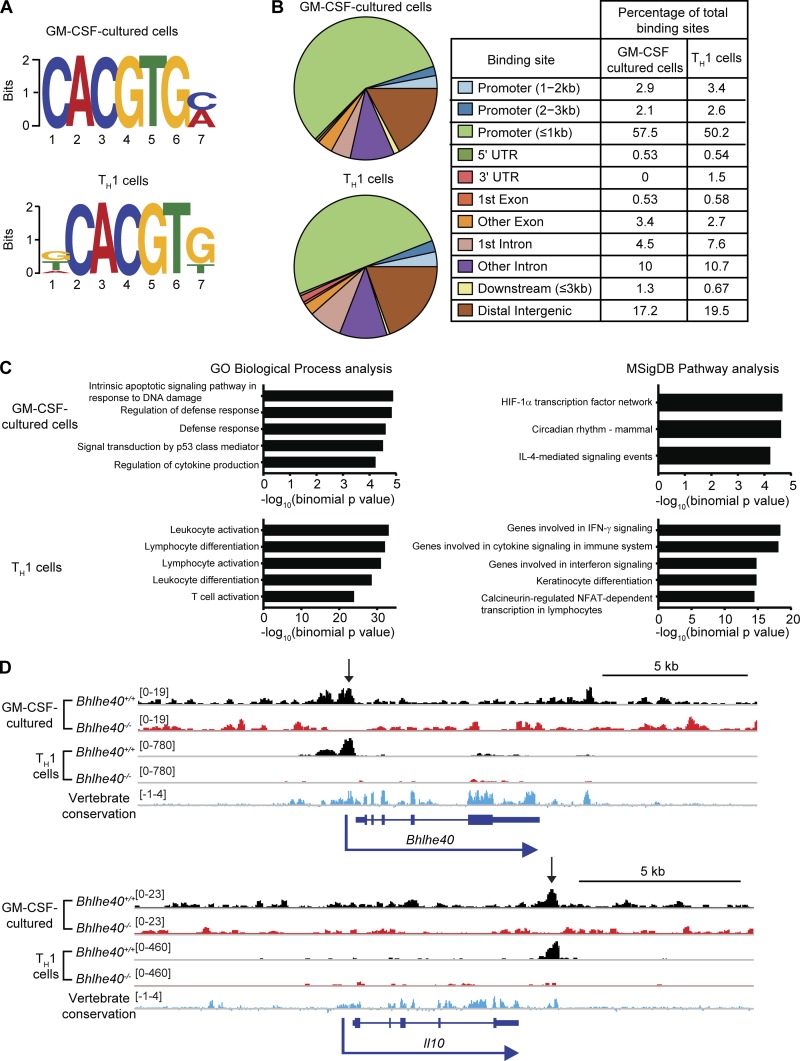
Figure 6.
Bhlhe40 directly binds the Il10 locus in myeloid and lymphoid cells. (A–D) Bone marrow cells were differentiated in the presence of GM-CSF and stimulated with heat-killed Mtb for 4 h. CD4+ T cells were TH1 polarized in vitro for 4 d. DNA was immunoprecipitated using anti-Bhlhe40 antibody and sequenced. (A) Sequence motifs present within DNA bound by Bhlhe40 were analyzed by MEME-ChIP. (B) Bhlhe40 binding sites were annotated using ChIPseeker (3.5). (C) Functions of cis-regulatory regions were predicted for Bhlhe40 binding site data using GREAT. The top five most highly enriched gene sets with minimum region-based fold enrichment of 2 and binomial and hypergeometric false discovery rates of ≤0.05 in the Gene Ontology (GO) Biological Process and MSigDB Pathway gene sets are displayed for each dataset (in GM-CSF–cultured cells, only three MSigDB Pathway gene sets met these criteria). NFAT, nuclear factor of activated T cells. (D) ChIP-seq binding tracks for Bhlhe40 at the Bhlhe40 and Il10 loci in Bhlhe40+/+ and Bhlhe40−/− myeloid and lymphoid cells. Vertebrate conservation of each genomic region is displayed in blue, and peaks identified by MACS are indicated by arrows. Bracketed numbers indicate the trace height range. Data are from a single experiment.