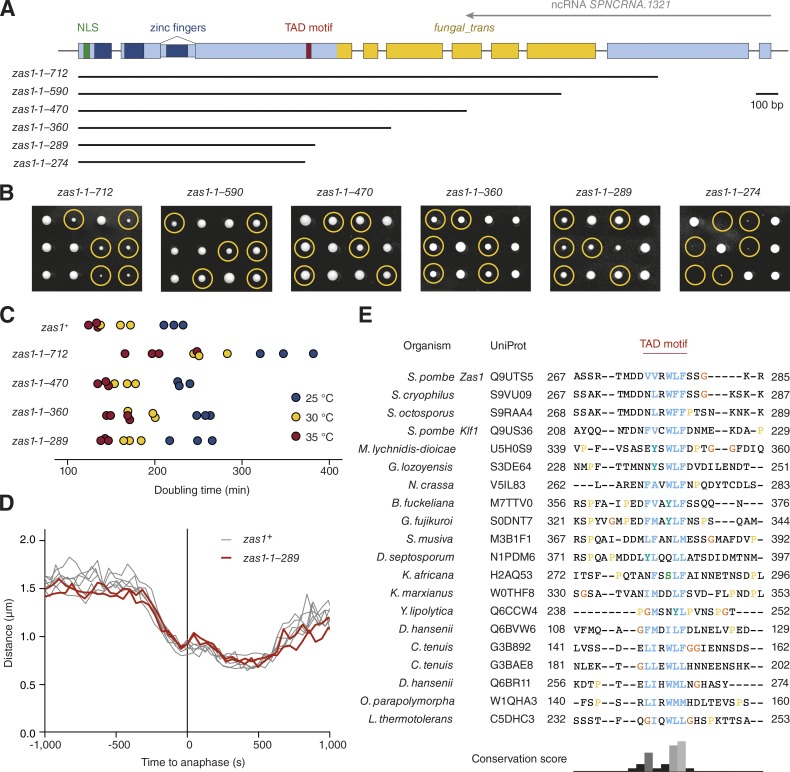
Figure 6.
Identification of a conserved TAD motif in Zas1. (A) Schematic overview of zas1 truncation constructs. Numbers indicate the amino acid residue range in each construct. (B) Tetrad analysis of heterozygous zas1-1–x/zas1+ diploid fission yeast strains (C4006, C4445, C4007, C4036, C4087, C4035) after 5 d at 25°C. Circles identify the position of spores bearing the zas1-1–x gene. (C) Doubling times of cells expressing truncated versions of the zas1 gene (C3974, C4026, C4025, C4037, C4047) in rich liquid medium (YE5S) measured in three independent experiments at 25°C, 30°C, and 35°C. (D) Condensation curves of cells expressing the truncated zas1-1–289 (red, C4094, n = 2 independent experiments) and wild-type zas1+ control strains (black lines, n = 6 independent experiments from Fig. 1). (E) Top hits of a protein sequence alignment of Zas1 homologues in different species from a PSI-BLAST search (see supplementary alignment file). Numbers indicate start and end residue numbers of the sequences in the alignment.