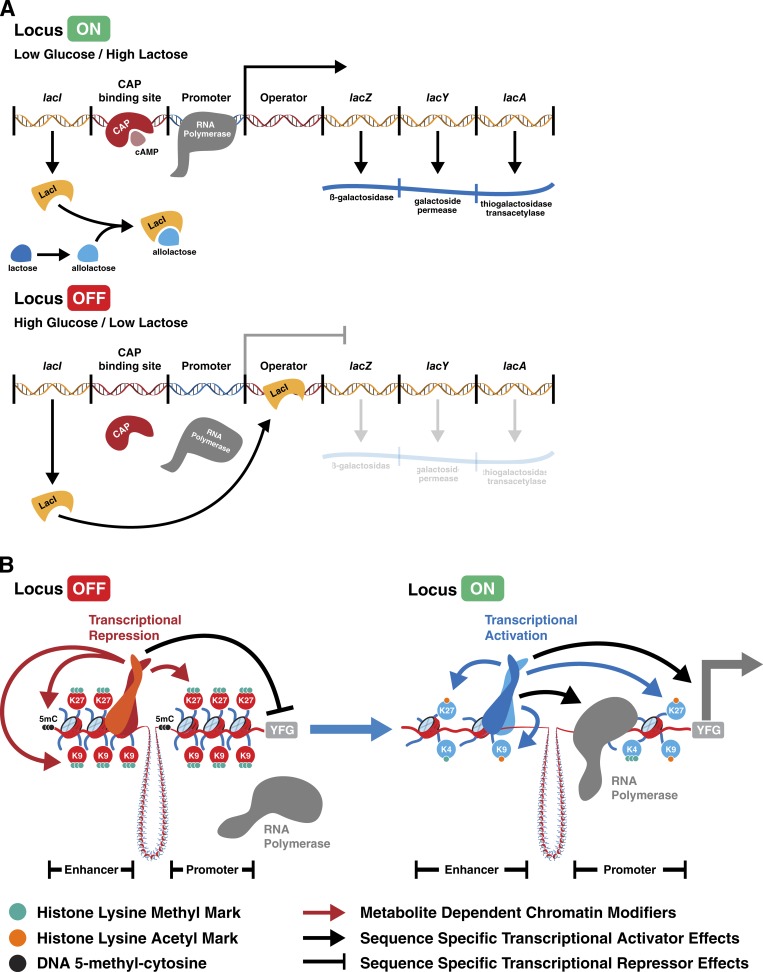
Figure 1.
Paradigms of metabolic regulation of gene expression. (A) Summarized model of the E. coli lac operon as outlined by Jacob and Monod (1961). In low glucose/high lactose conditions, the lac repressor (LacI) binds allolactose and RNA polymerase is able to activate transcription of genes required for lactose metabolism. Conversely, in high glucose/low lactose conditions, LacI is not bound to allolactose and can bind to the operator sequence, repressing the ability of RNA polymerase to transcribe operon genes. CAP, catabolite activator protein. (B) Schematic representation of how sequence-specific DNA binding proteins recruit chromatin modifying enzymes that serve to deposit inhibitory (left) or activating (right) marks. In this model, transcription factors recruit local chromatin modifying enzymes. YFG, your favorite gene; 5mC, 5-methyl-cytosine; K9, histone H3 lysine 9; K27, histone H3 lysine 27; K4, histone H3 lysine 4.