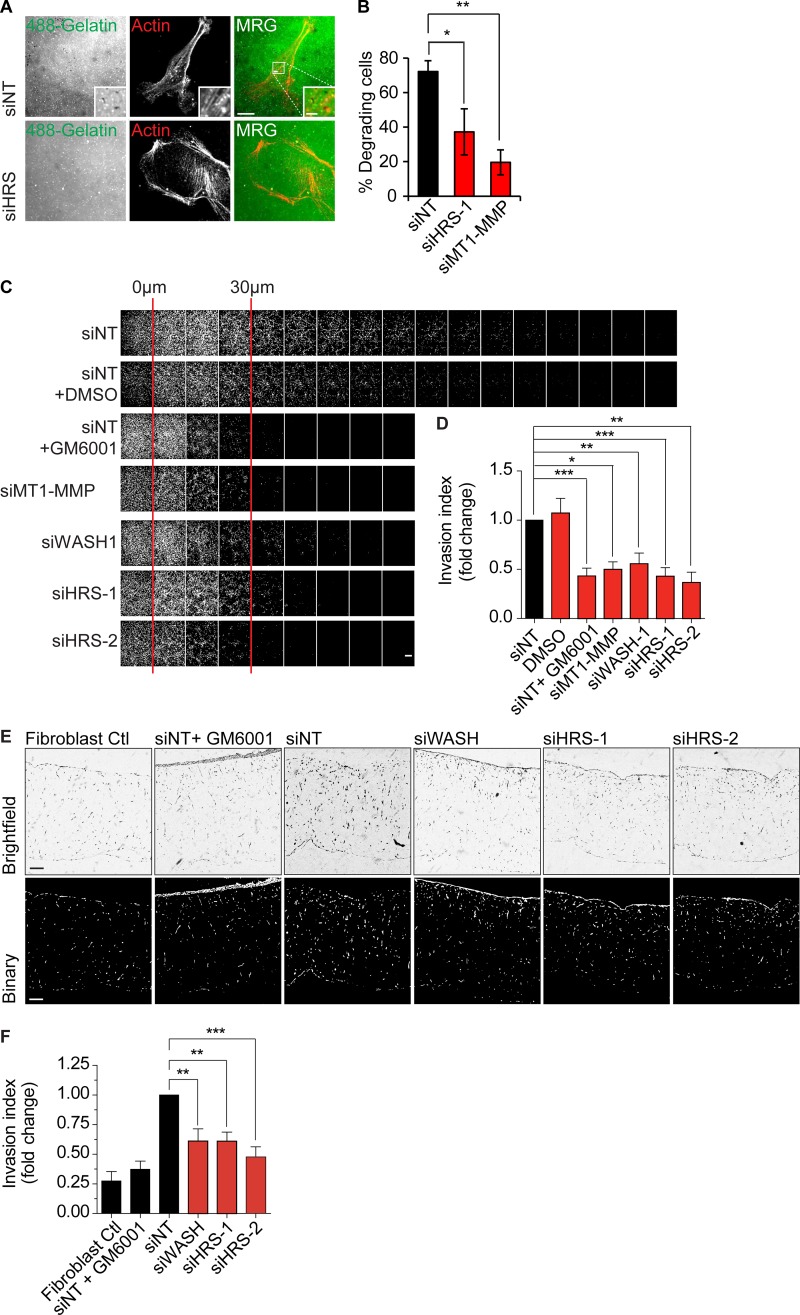
Figure 8.
HRS and WASH axis are required for matrix degradation and breast cancer cell invasion. (A) MDA–MB-231 cells were treated with siRNA for 120 h before being seeded onto 488-gelatin–coated coverslips for 16 h. MRG, merge. (B) Percentage of degrading cells (n = 3 individual experiments; approximately >60 cells total). Images taken from a single slice. (C) MDA–MB-231 cells were treated with siRNA for 96 h before 8 × 104 cells seeded for an inverted invasion assay on a gel composed of Matrigel/fibronectin followed by culture for 5 d. Images were taken every 10 µm and analyzed using ImageJ. (D) Relative invasion index over 30 µm (n ≥ 3). 20 ng/ml EGF was used as a chemoattractant. GM6001 (5 µM), DMSO (1:1,000), and gels without cells were used as invasion, vehicle, and fibroblast background controls, respectively. (E) Organotypic raft culture. MDA–MB-231 cells were treated with siRNA for 96 h before 7 × 105 cells seeded on fibroblast remodeled collagen gels and allowed to adhere for 3 d. Gels were then cultured at the liquid–air interface for a further 7 d. Gels were fixed, embedded, hematoxylin and eosin–stained, and sectioned to determine invasion index. Bars: (main images) 10 µm; (insets) 2 µm. (F) Quantification of MDA–MB-231 cell invasion relative to siNT control (n = 4 independent experiments). Error bars indicate SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Statistical analysis, one-way ANOVA for all comparisons with Dunnett’s post hoc test.