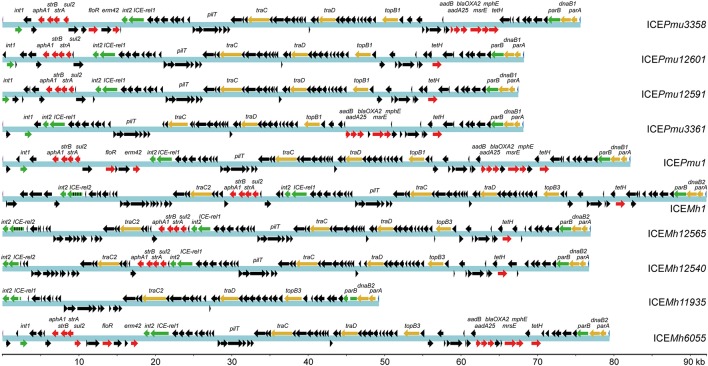
Figure 2.
Content of ICEs from the Pasteurellaceae strains determined by whole genome sequencing. The previously reported ICEPmu1 of P. multocida strain 36950 (Michael et al., 2012b) and ICEMh1 from M. haemolytica strain 42548 (Eidam et al., 2015) are included for comparison. The ICEs are annotated here according to the strains containing them, and their sizes can be read from the kilobase pair (kb) scale at the bottom. Color coding of the genes as in Figure 1 with the core genes targeted in the multiplex assay (green), other essential core genes (yellow), antibiotic resistance genes (red) plus additional open reading frames (black). The chromosomal site of ICE insertion left a disrupted tRNALeu gene at both ends of the sequence, which is replaced by in all cases by an intact copy of the tRNALeu gene located after parA (far right, in this sequence orientation). The multiplex assay was design to give a positive signal for ICE-rel1 but not for the ICE-rel2 gene (green/black stripes) that is evident in some M. haemolytica ICEs. The sensitive strain Pmu4407 contains no ICE genes. Strain Mh11935 also completely lacks resistance genes but contains numerous ICE core region genes, although key genes for ICE transfer and maintenance are missing. The ICE-rel1 gene in ICEMh11935 is truncated and presumably inactive, despite giving a positive multiplex PCR signal. The parB gene of ICEMh11935 is also truncated, and lacks one of the priming regions needed for a PCR signal. All of the four targeted genes int1, int2, ICE-rel1, and parB were 100% conserved in ICEPmu3358, ICEPmu3361, ICEPmu12591, and ICEPmu12601 of the P. multocida strains. Accession codes for the whole genome sequences are listed in Supplementary Table S1.