Figure 3.
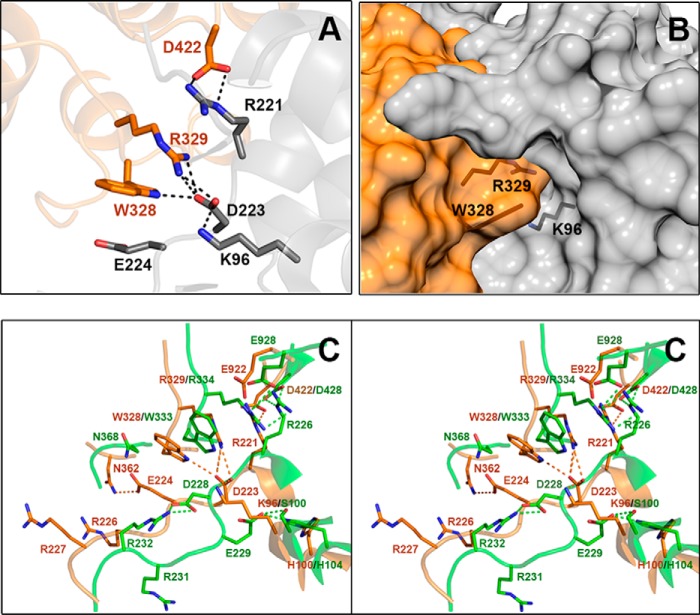
Region of FtPEPC-C4 equivalent to the allosteric-site for neutral amino acids in ZmPEPC-C4. A, cartoon representation showing that Lys-96 (equivalent to Ser-100 of ZmPEPC-C4) interacts with Asp-223 (equivalent to Asp-228 of ZmPEPC-C4), which in turn is hydrogen-bonded to Arg-329 (equivalent to Arg-334 of ZmPEPC-C4) and Trp-328 (equivalent to Trp-333 of ZmPEPC-C4). Amino acid side chains are depicted as sticks with carbon atoms in orange or gray (depending on the monomer of the dimeric unit), oxygen atoms in red, and nitrogen atoms in blue. B, surface representation of the monomer–monomer interface of the FtPEPC-C4 dimer, showing that the allosteric site for neutral amino acids does not exist because the position of the above mentioned residues does not leave a cavity between the two monomers. The side chains of relevant protein residues are shown as sticks with oxygen atoms in red, nitrogen in blue, and carbons in black. Of the three reported crystal structures of FtPEPC-C4, the one shown here is Protein Data Bank code 3ZGE. C, differences in the conformation of critical residues of the allosteric site for amino acids between the 5VYJ ZmPEPC-C4 (green carbon atoms) and FtPEPC-C4 3ZGE (orange carbon atoms) crystal structures. Hydrogen bonds are depicted as dashed lines, black in A and green (maize isozyme) or orange (Flaveria isozyme) in C; cutoff is 3.0 Å. A and C were generated using PyMOL, and B was generated using the UCSF Chimera package (52).