Abstract
Preeclampsia is the most common clinical disorder in pregnancy and might result from disordered uterine environments caused by epigenetic modifications, including deregulation of DNA methylation/demethylation. Recent research has indicated that 5-hydroxymethylcytosine (5hmC), a DNA base derived from 5-methylcytosine (5mC) via oxidation by ten-eleven translocation (TET) enzymes, is involved in DNA methylation–related plasticity. Here, we report that TET2 expression and 5hmC abundance are significantly altered in the placentas from preeclampsia patients. shRNA-mediated TET2 knockdown (shTET2) reduced trophoblast migration and invasion when cultured in Matrigel. Both real-time PCR of matrix metalloproteinase (MMP)–related transcripts and a human angiogenesis antibody array indicated that TET2 knockdown in trophoblasts inhibits the expression of MMP transcript, of which MMP9 represented one of the most significant TET2 downstream targets. Using an established shTET2 HTR-8/SVneo cell model, we further confirmed alterations of 5hmC levels and MMP9 expression at both mRNA and protein levels. In particular, we found that TET2 bound to and removed 5mC modifications at the MMP9 promoter region. Interestingly, in TET2 knockdown cells, both MMP9 expression and the compromised trophoblast phenotype could be rescued by vitamin C, an activator of TET enzyme activity. Finally, TET2 expression correlated with MMP9 levels in placenta samples from the preeclampsia patients, indicating that TET2 deregulation is critically involved in the pathogenesis of preeclampsia through down-regulation of MMP9 expression. Our findings highlight a critical role of TET2 in regulating trophoblast cell migration through demethylation at the MMP9 promoter, and suggest that down-regulation of the TET2–MMP9–mediated pathway contributes to preeclampsia pathogenesis.
Keywords: epigenetics, pregnancy, placenta, metalloprotease, DNA demethylation, preeclampsia, pregnancy complication, TET2, trophoblast, MMP9, methylation
Introduction
Preeclampsia (PE)4, a pregnancy-related disease with multiorgan involvement, is primarily defined by the occurrence of new-onset hypertension plus proteinuria after 20 weeks of gestation (1). It is complicated in 2–8% of all pregnancies, with an estimated 46,000 PE-related deaths in 2015 worldwide (2–4). Furthermore, the incidence of perinatal diseases, such as heart disease, is significantly increased with PE; thus, prediction and prevention of PE are urgently needed considering the disease risks (5, 6). Angiogenic and anti-angiogenic factors are secreted from trophoblast cells, and imbalanced production/secretion is regarded as a major etiological factor for preeclampsia (7, 8). Currently the sFlt-1/PlGF ratio has gained much attention as a useful biomarker because it has a very high predictive value for maternal and/or fetal PE-related adverse events. The so-called angiogenic PE (defined as a sFlt-1/PlGF ratio >85) is regarded as the most harmful condition (9). However, the variable phenotypes of PE indicate that other pathological factors also contribute to its incidence.
Epigenetic changes in the placenta, including noncoding RNA expression and DNA methylation modification are now regarded as major mechanisms in the placentation process and their deregulation contributes significantly to placenta-related diseases like PE (8). Trophoblast methylation/demethylation is a major mechanism for differentiation and development. Although it is generally accepted that the methylation process is inhibitory during preimplantation embryonic development, it has been recently shown that active demethylation processes occur after implantation (10, 11). Deregulation of the demethylation process could cause pregnancy-related diseases like PE. In fact, PE has recently been associated with aberrant methylation of tumor suppressor genes in the placenta (12). 5hmC is considered an intermediate in an active demethylation process catalyzed by the ten-eleven translocation (TET) family, which comprises three members, TET1 to TET3, that have combinatory roles in different developmental process as well as carcinogenesis (13). Recent data have shown that TET is involved in 5hmC-mediated alteration of the transposon-mediated activity associated with exposure to adverse in utero environments, and both TET1 and TET3 are down-regulated in PE placenta, indicating an important role in PE pathogenesis (14). Compared with TET1 and TET3, TET2 is unique as it lacks the CXXC domain and requires IDAX to recruit to target genes and regulate its stability. Specifically, a recent study shows that TET2 is also critical for hematopoiesis and that TET2 mutation is related to myeloid tumorigenesis (15, 16). Whether TET2 plays critical roles in the placentation process and the underlying mechanisms remain unclear.
During placenta development, cytotrophoblast proliferation, invasion, and syncytiotrophoblast formation are orchestrated steps critical for normal pregnancy (17). Trophoblast migration is an important aspect of implantation and placenta development, as shallow implantation is generally regarded as a major mechanism of PE (7). The normal trophoblast invasion process is similar in mechanism to tumor metastasis. Matrix metalloproteinases (MMPs) are well-known secreted proteins important for both normal embryonic development and tumor metastasis. Mounting evidence, including ours, shows that they are also important for trophoblast invasion, and thus, MMPs are drug targets for prevention of PE (18, 19). Among more than 20 members of the MMP family, MMP9 is noteworthy because knockout of MMP9 induces the PE phenotype in mice (20). Reduced MMP9 protein secretion is also observed in PE patients, thus supporting a critical role for MMP9 in trophoblast migration/invasion (21, 22).
In the current study, we have shown that TET2 protein can regulate MMP9 expression and trophoblast invasion. We further delineated the detailed mechanism of how reduced TET2 expression leads to compromised trophoblast invasion, and revealed a critical regulatory pathway involved in PE development.
Results
Decrease in 5hmC is associated with a substantial reduction of TET2 gene expression in PE placenta
The role of TET2 protein in the placenta is yet to be determined. We measured the expression of TET2 protein and the levels of 5mC and 5hmC at genomic DNA in human placenta. In the dot-blot assay, the level of 5hmC/5mC in the PE placenta was lower than the control (Fig. 1A). Conversion of 5mC to 5hmC was mediated by TET enzymes. TET2 mRNA expression measured by real-time PCR was reduced by 35.7% in the PE group compared with control group (Fig. 1B, p < 0.05). TET2 protein expression was significantly reduced by Western blotting in the PE group compared with control (Fig. 1C, p < 0.001). Furthermore, immunohistochemistry analysis showed that TET2 protein staining was localized in the cytotrophoblast as well as in the syncytiotrophoblast, and that staining intensity was reduced in the PE patient (Fig. 1D). The 5hmC level was also reduced in placental sections as indicated by immunohistochemistry (Fig. 1D). Together, consistent with the reduced TET2 protein expression, 5hmC level was significantly reduced in placenta samples from PE patients compared with control.
Figure 1.
Reduced 5hmC was associated with substantial reduction of TET2 expression in the PE placenta. A, dot-blot assay result showed that the 5hmC was down-regulated at genomic DNA in PE placentas (n = 13) compared with controls (n = 11). Methylene blue was used as a loading control. B, real-time PCR results show that TET2 gene expression was decreased in PE placentas (n = 13) compared with controls (n = 11); GAPDH was used as an endogenous control. C, Western blotting results show that TET2 protein expression was reduced in PE placentas (n = 13) compared with controls (n = 11). Tubulin was used as an endogenous control. D, immunohistochemical staining of TET2, and 5hmC in PE placental sections (n = 5) and controls (n = 5). TET2 was mainly expressed in cytotrophoblast cells in the placental villi. Scale bar, 100 μm. The n means the number of samples in each group. The histogram shows statistical results of biological replicates. The data are presented as the mean ± S.D. **, p < 0.01; *, p < 0.05.
TET2 plays important roles in human trophoblast cell proliferation, invasion, and migration by regulating MMP9 gene expression
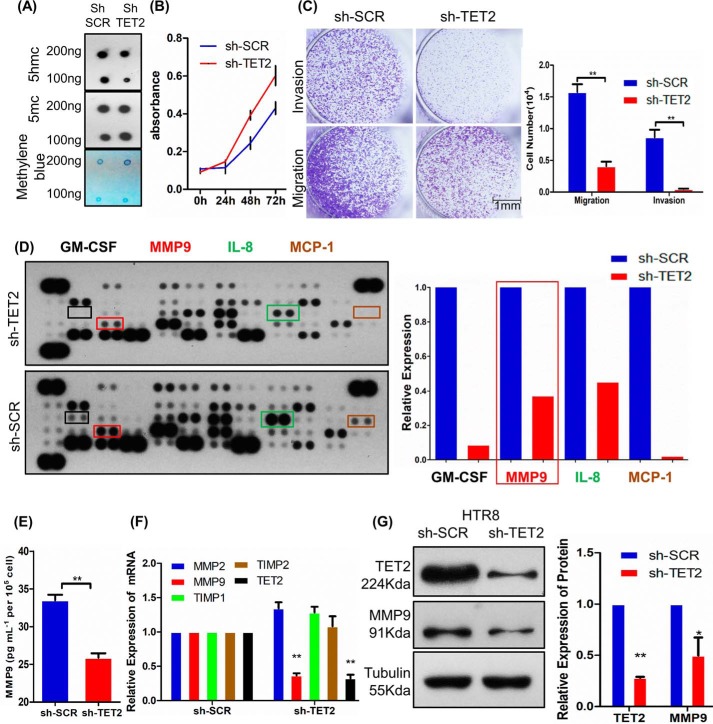
As TET2 is expressed in trophoblast cells, we investigated whether TET2 could affect trophoblast cell behavior using cell model. The 5hmC level was significantly reduced after using specific shRNAs to reduce TET2 expression at genomic DNA in HTR-8/SVneo cell (shTET2) compared with control cells (shSCR) by dot-blot assay (Fig. 2A). We then evaluated the proliferation, invasion, and migration ability of HTR-8/SVneo cell after knockdown TET2. The Cell Counting Kit–8 assay showed that shTET2 can induce proliferation (Fig. 2B). Furthermore, the migration and invasion ability of HTR-8/SVneo cell were assessed using Transwell assays. The results showed that shTET2 caused significant inhibition of cell migration and invasion (Fig. 2C).
Figure 2.
TET2 plays important roles in human trophoblast cell proliferation, invasion, and migration. A, the level of 5hmC was down-regulated after lentivirus-mediated knockdown TET2 in HTR-8/SVneo cell. Methylene blue was used as a loading control. B, cell proliferation as measured by cell numbers at different time points. C, the knockdown of TET2 inhibits invasion and migration ability in HTR-8/SVneo cell. Scale bar, 1 mm. D, angiogenesis-related antibody array indicates that the secretion of proteins, such as GM-CSF, MMP9, IL-8, and MCP-1, was decreased. E, ELISA confirmed that MMP9 levels are significantly reduced in shTET2 compared with shSCR. F, real-time PCR shows that MMP9 was decreased in shTET2 compared with shSCR. GAPDH was used as an endogenous control. G, Western blotting shows that MMP9 protein was reduced in shTET2 compared with shSCR. Tubulin was used as an endogenous control. All experiments were repeated a minimum of three times, and representative data are shown. The data are presented as the mean ± S.D. **, p < 0.01; *, p < 0.05.
To investigate the relationship between TET2 and angiogenesis ability in HTR-8/SVneo cell, an angiogenesis antibody array was performed to determine expression level of angiogenesis-related proteins in conditioned media derived from shSCR and shTET2 cell. The results showed that the expression levels of GM-CSF, MMP9, IL-8, and MCP-1 were significantly decreased in shTET2 supernatant compared with shSCR supernatant (Fig. 2D). The ELISA result further confirmed that MMP9 expression level was significantly decreased in shTET2 supernatant compared with shSCR supernatant, which was consistent with results of angiogenesis antibody array (Fig. 2E).
It is well known that placental trophoblast invasion is significant for maintenance of normal pregnancy. This phenomenon requires balanced expression between MMPs and their inhibitors, TIMPs. Thus, we analyzed the mRNA expression level of major MMPs, such as MMP2, MMP9, and their respective inhibitors in HTR-8/SVneo cell. Real-time PCR results showed that MMP9 expression was decreased by 63.4% in shTET2 (Fig. 2F, p < 0.01). Furthermore, MMP9 protein expression also significantly decreased by Western blotting in shTET2 compared with shSCR (Fig. 2G, p < 0.05).
TET2 could demethylate CpG sites in the MMP9 promoter and regulate promoter activity
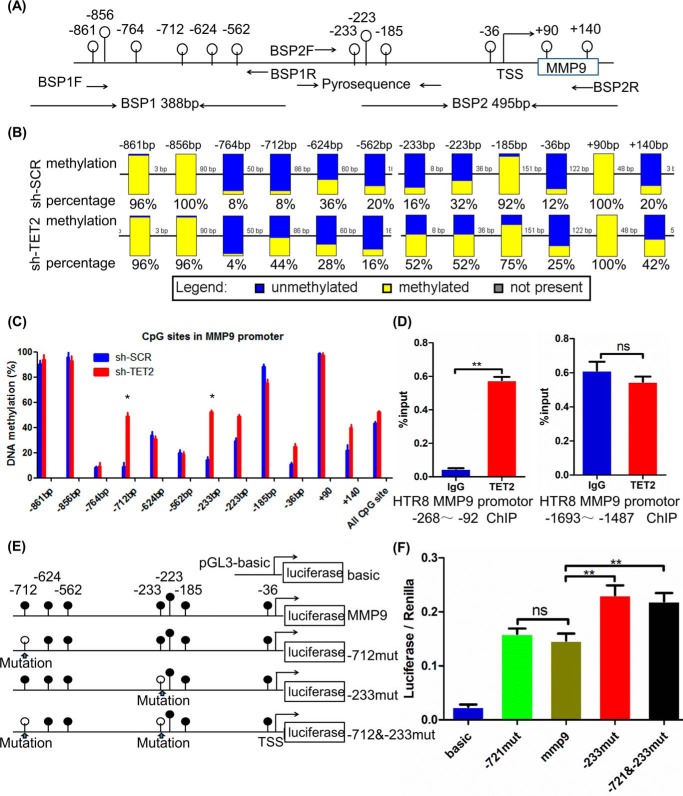
To investigate the relationship between TET2 and MMP9 expression, the methylation levels of the MMP9 promoter region were determined by bisulfite sequencing PCR (BSP) analysis. The promoter sequence of MMP9 from −761 bp to the transcription start site has the strongest transcriptional activity (23). Therefore, this region was chosen for the detailed methylation analysis. The MMP9 promoter region evaluated in this study is illustrated schematically in Fig. 3A. The BSP results demonstrated that overall DNA methylation percentage of the MMP9 promoter was relatively high in the control cells (15%). However, the DNA methylation percentage for each particular site was variable. Among them, eight CpG sites (−764, −712, −624, −562, −233, −223, −36, and +140 bp) had a methylation percentage below 50% (Fig. 3B), indicating that demethylation prevailed in these sites under normal conditions. However, the methylation percentages increased significantly at the −712 bp and −233 bp sites in shTET2 (Fig. 3C, p < 0.05).
Figure 3.
TET2 demethylates CpG sites in the MMP9 promoter and regulates promoter activity. A, schematic diagram of the MMP9 promoter. The white circles above the solid line represent the CpG in the promoter. The position and orientation of the bisulfite sequencing and pyrosequencing primers are indicated by black arrows. B, DNA methylation percentages at specific CpG sites are plotted for each individual DNA template. Each datum was calculated from the sequencing results of 25 independent clones. C, DNA methylation percentages at specific CpG sites are plotted for shTET2 and shSCR cells. D, ChIP analysis results showed that the TET2 level was increased compared with IgG in MMP9 promotor −268 to −92 bp. E, line drawings showing various methylation states. The white circles represent CG mutated to AT, similar to demethylated CpG sites, and black circles represent methylated CpG sites. F, each of the promoter sequences described above was inserted into the pGL3-Basic vector for the Dual-Luciferase reporter assay. Transcriptional activities of promoters with various methylation states are shown. All experiments were repeated a minimum of three times, and representative data are shown. The data are presented as the mean ± S.D. **, p < 0.01; *, p < 0.05.
To estimate whether the TET2 protein binds to MMP9 promoter regions, we quantified the level of TET2 enrichment in the MMP9 promoter at the −268 to −92 region in HTR-8/SVneo cells by chromatin immunoprecipitation (ChIP) assay. The ChIP analysis results showed that the TET2 level was increased by 14.5-fold compared with IgG (Fig. 3D, p < 0.01). Briefly, TET2 could bind and demethylate CpG sites in the MMP9 promoter region, especially at the −233 bp site. Therefore, when the MMP9 promoter activities with various methylation profiles were evaluated by the Dual-Luciferase reporter assay, the results showed that the −712mut sequence activity was not significantly different compared with normal MMP9 promoter, whereas the −233mut and the −712 and −233mut sequence activity was increased by 36 and 33%, respectively, compared with normal MMP9 promoter (Fig. 3F, p < 0.01). Therefore, we concluded that TET2 could demethylate CpG sites, especially at the −233 bp site in the MMP9 promoter and thus control promoter activity, whereas other CpG sites may play supportive roles in this regulatory mechanism. In conclusion, these results suggest that TET2 could regulate the proliferation, migration, and invasion of HTR-8/SVneo cells through its target, MMP9 in trophoblasts, which could play an essential role in PE pathophysiology.
Re-expression of TET2 catalytic domain (Tet2-CD) could partly rescue the migration and invasion in shTET2 cell
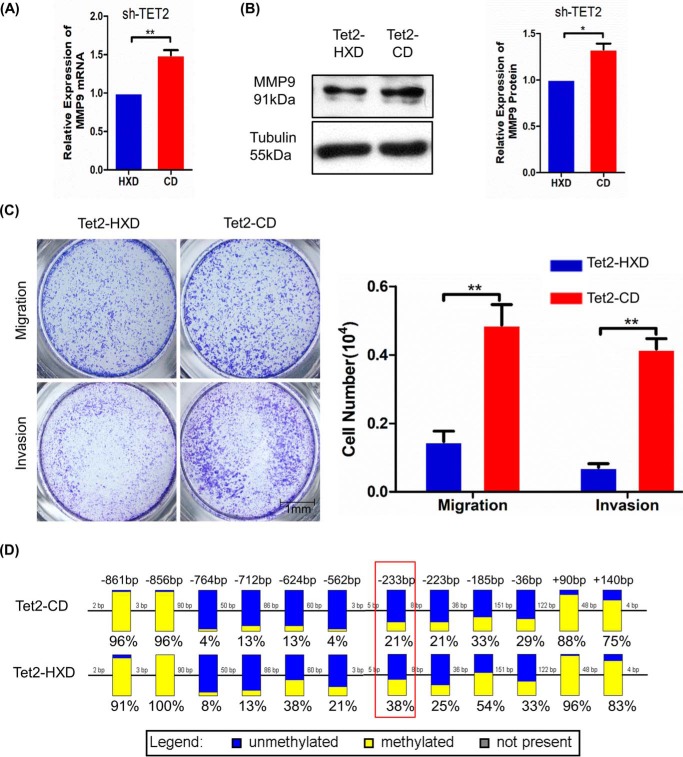
To further study whether TET2 enzyme activity was required for MMP9 expression. We transfected, respectively, an expressed plasmid of TET2 catalytic domain (Tet2-CD) and catalytically inactive version of Tet2-CD (Tet2-HXD) in shTET2 cell (24). The real-time PCR results showed that transfected Tet2-CD could modestly, but significantly, enhance MMP9 mRNA expression compared with Tet2-HXD (Fig. 4A), and protein level shows same trend in the Western blot results (Fig. 4B). We measured the migration and invasion ability of shTET2 cell after re-expression TET2. The Transwell assay showed that re-expression of Tet2-CD could enhance the migration and invasion in shTET2 cell compared with Tet2-HXD (Fig. 4C). We next tested the level of methylation of MMP9 promotor. BSP results showed that overall DNA methylation level of the MMP9 promoter was relatively low in the transfected Tet2-CD cell; the single CpG site methylation percentages decreased significantly at the −233 bp sites in the transfected Tet2-CD cell (Fig. 4D). Therefore, re-expression of TET2 could rescue the MMP9 expression through its enzyme activity.
Figure 4.
Re-expression of TET2 catalytic domain (Tet2-CD) could partly rescue the migration and invasion in shTET2 cell. A, real-time PCR shows that MMP9 was increased in shTET2 cell after transfected Tet2-CD compared with Tet2-HXD. GAPDH was used as an endogenous control. B, Western blotting shows that MMP9 protein was increased in shTET2 cell after transfected Tet2-CD compared with Tet2-HXD. Tubulin was used as an endogenous control. C, the transfection of Tet2-CD rescued invasion and migration ability in shTET2 cell. Scale bar, 1 mm. D, DNA methylation percentages of MMP9 promoter at specific CpG sites are plotted for each individual DNA template. Each datum was calculated from the sequencing results of 24 independent clones. All experiments were repeated a minimum of three times, and representative data are shown. The data are presented as the mean ± S.D. **, p < 0.01; *, p < 0.05.
Decrease of MMP9 correlates with DNA demethylation on MMP9 promoter in PE placenta
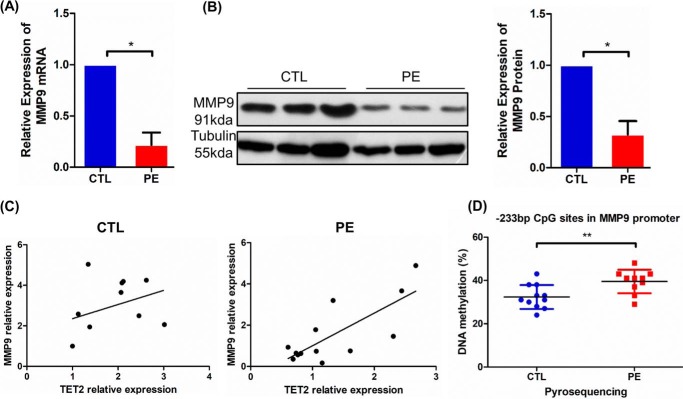
Based on the above experiment, we investigated the expression of MMP9 in PE and control placenta by real-time PCR and Western blotting. MMP9 mRNA expression measured by real-time PCR was reduced by 46% in placenta from the PE group compared with control group (Fig. 5A, p < 0.05). The placental protein expression of MMP9 was clearly reduced by Western blotting in the PE group compared with controls (Fig. 5B, p < 0.05). Pearson's correlation assay demonstrated that TET2 expression was positively correlated with the MMP9 mRNA level in PE placenta (Fig. 5C, r2 = 0.57, p = 0.0043), but not with that in the control (Fig. 5C, r2 = 0.13, p = 0.281). Furthermore, we examined the status of MMP9 promoter methylation by pyrosequencing in PE and control placenta. The results indicated that the −233 bp site of the MMP9 promoter shows higher methylation levels in PE (Fig. 5D, p < 0.01). Taken together, these findings reveal that MMP9 methylation and expression levels were changed in PE placenta compared with controls.
Figure 5.
Decrease of MMP9 correlated with DNA demethylation on MMP9 promoter in PE placenta. A, real-time PCR results showed that MMP9 was decreased in PE placentas (n = 13) compared with that in controls (n = 11). GAPDH was used as an endogenous control. B, MMP9 expression was significantly reduced in PE placentas (n = 13) compared with that in controls (n = 11) by Western blotting. Tubulin was used as an endogenous control. C, Pearson's correlation assay demonstrated that TET2 expression was positively correlated with MMP9 mRNA levels in the PE placenta (n = 13, r2 = 0.57, p = 0.0043), but not with that in the control (n = 11, r2 = 0.13, p = 0.281). GAPDH was used as an endogenous control. D, pyrosequencing results indicated that the −233 bp site of the MMP9 promoter was hypermethylated in the PE placentas (n = 10) compared with controls (n = 11). The n means the number of samples in each group. The histogram shows statistical results of biological replicates. The data are presented as the mean ± S.D. **, p < 0.01; *, p < 0.05.
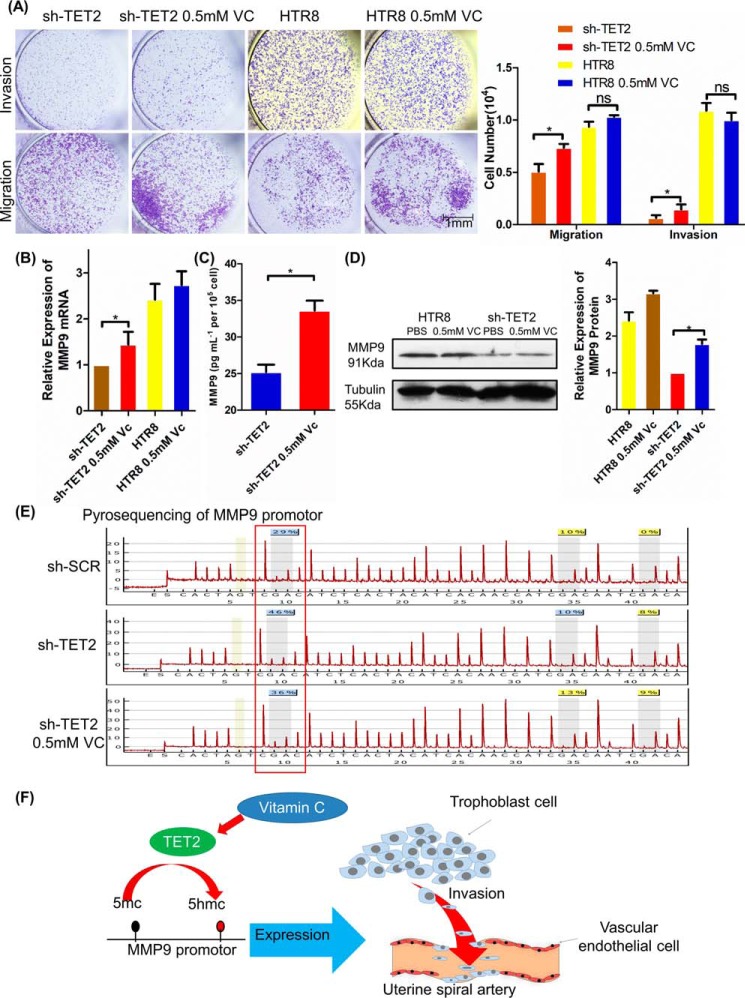
Vitamin C treatment restores TET2 activity and MMP9 gene expression in HTR-8/SVneo cells
To investigate whether TET2 knockdown could be rescued by the enzymatic activity of vitamin C (VC), which enhances TET2 catalytic activity, we measured the invasion and migration ability of HTR-8/SVneo cells after VC treatment. The Transwell assay showed that VC treatment remarkably induced cell migration and invasion in shTET2 and in HTR-8/SVneo cells (Fig. 6A). We next assessed whether MMP9 expression could be rescued by VC treatment. The real-time PCR results showed that MMP9 mRNA level was rescued after VC treatment in shTET2 compared with shTET2 (Fig. 6B, p < 0.05). The ELISA results further confirmed that MMP9 expression also significantly increased in VC treatment shTET2 supernatant compared with shTET2 supernatant (Fig. 6C, p < 0.05). Next, the Western blot result showed that MMP9 protein expression clearly increased by VC treatment in shTET2 compared with shTET2 (Fig. 6D, p < 0.05). Furthermore, we examined the MMP9 promoter methylation status by pyrosequencing after VC treatment in shTET2. The result demonstrated that the hypermethylated −233 bp site of the MMP9 promoter could be rescued, at least partially, by VC treatment in shTET2 (Fig. 6E).
Figure 6.
Vitamin C treatment restored TET2 activity and MMP9 gene expression in HTR-8/SVneo cells. A, VC treatment up-regulated TET2 enzyme activity and rescued cell invasion and migration in shTET2 cells and in normal HTR-8/SVneo cells. Scale bar, 1 mm. B, real-time PCR showed that MMP9 was increased after VC treatment in cells. GAPDH was used as an endogenous control. C, the ELISA results further confirmed that MMP9 expression significantly increased in VC treatment shTET2 supernatants compared with shTET2. D, Western blotting showed that MMP9 expression was induced after VC treatment in cells. Tubulin was used as an endogenous control. E, pyrosequencing results indicated that the −233 bp site of the MMP9 promoter was demethylated after VC treatment in shTET2 cells. F, the proposed model for the regulation of demethylation levels on the MMP9 promoter in trophoblasts by TET2: TET2 binds to MMP9 promoter and down-regulates its methylation; MMP9 expression and trophoblastic invasion is therefore up-regulated. VC could rescue the effect of TET2 on MMP9 expression by up-regulating TET2 enzyme activity. All experiments were repeated a minimum of three times, and representative data are shown. The data are presented as the mean ± S.D. **, p < 0.01; *, p < 0.05.
In brief, our result showed that TET2 bound to MMP9 promoter and was led to its hypomethylation. Therefore, the expression of MMP9 was increased, which promotes the invasion ability of trophoblast cell. On the other hand, VC could rescue the effect of TET2 on MMP9 by up-regulating TET2 enzyme activity (Fig. 6F).
Discussion
The current study highlights the critical role of TET2 in regulating trophoblast cell invasion through regulating MMP9 promoter demethylation, especially at the −233 CpG site in the promoter region. Furthermore, it shows that PE placentas have aberrant methylation levels in the MMP9 promoter region, indicating that TET2-mediated hypermethylation could be responsible for altered MMP9 expression; thus our study revealed a critical pathway involved in the placentation process, and its deregulation results in PE pathogenesis.
The MMP9 promoter has been extensively studied and a CpG island has been found in this promoter region (23, 25–27). However, our study indicates that another CpG region is involved in the hypermethylation process and is critical for MMP9 transcription. Furthermore, we show that TET2 could bind to specific sites on the MMP9 promoter region, such as the −233 and −712 CpG sites; control the demethylation process; and affect MMP9 promoter transcriptional activity. In our study, we found that the −233 bp region of the MMP9 promoter is important for demethylation, and ChIP assay further demonstrated that TET2 could bind to the MMP9 promoter CpG site and demethylate the CpG site. TET2 has been shown to interact with DNA methyltransferase in the promoter region and dynamically regulate oxidative stress-related cancer cell responses (11, 28). Other histone modifications also play roles in methylation regulation of MMP9 promoter (29, 30). Furthermore, acetylation of TET2 could increase TET enzyme activity and is therefore critical for the TET-mediated demethylation process (28). Thus, the exact mechanism of how TET2 controls MMP9 expression requires further investigation.
Epigenetic modifications including DNA methylation, RNA regulation, and imprinting are critically involved in placenta development; adverse in utero environments such as ART (Assisted Reproductive Technology) manipulation and infection have been shown to affect placenta development by regulating methylation levels (31, 32). Our result showing reduced TET2 expression indicates that TET2-mediated hypermethylation plays an important role in the placentation process. Although it has been shown that TET1 and TET3 are reduced in PE placenta, this study is the first to show that the TET2–MMP9 pathway is also critically involved in PE pathogenesis. The exact mechanisms of how the upstream signals regulate TET2 expression need further investigation. For example, it has been shown that androgen could inhibit TET2 expression during prostate cancer progression (33). Recently abnormal androgen levels in the uterus have been related to the PE etiology in a murine model (34). Thus, whether abnormal androgen levels are involved in deregulating the TET2 expression in utero is also an interesting question.
Vitamin C is a well-known TET enzyme activator (35). A recent study shows that vitamin C is therapeutically effective for treating TET2-deficient leukemia by regulating stem cell proliferation (36, 37), indicating that vitamin C could also be potentially important for PE. Although the clinical benefits of vitamin C for PE treatment and prevention remain debatable, some studies show report prevention of DNA methylation associated with smoking after vitamin C/E intake, whereas others do not find significant effects of prenatal vitamin C intake for preventing PE (38, 39). Our study demonstrates that vitamin C could rescue the knockdown effects of TET2 as well as MMP9 expression, indicating that vitamin C could rescue some of effects in the placentation process. It is likely that vitamin C could stimulate other TET enzymes when TET2 is knocked down, therefore showing rescuing effects in our experiment. It also remains possible that the beneficial effects maybe more significant when the placenta is associated with adverse environments like oxidative stress. In the future, it would be important to check the TET2 levels/activity before evaluating the effects of vitamin C as a preventive strategy in future clinical study.
Methylation is now regarded as an important biomarker in PE (5), and genome-wide methylation deregulation has been shown as related to PE pathogenesis (40, 41). However, a study on the specific methylation at a single or several CpG sites could reveal more candidate biomarkers, which may represent more economic and straightforward biomarkers for clinic use (5). Our result shows that several candidate CpG sites, such as the −233 bp CpG site of MMP9 promoter, have differential methylation levels between PE and normal placenta, and further functional studies are warranted to further delineate the exact pathway elucidating the underlying mechanism of PE.
Materials and methods
Patients and tissue collection
Clinical samples were collected from women who visited the Obstetrics and Gynecology Department of West China Second University Hospital of Sichuan University in Chengdu, China. All study participants provided written informed consent. The study protocol was approved by the local ethics committee. The clinical diagnosis of PE is based on the standard as described in our previous study (8).
Placental tissue was obtained from severe PE cases (n = 13) and normal pregnancy controls (n = 11). Two groups of subjects were chosen and the placentas were cut after cesarean section on the maternal side, taking the tissue around the central villus area, rinsed with saline, quickly placed in liquid nitrogen, and then stored at −80 °C. One part of the fresh placenta was fixed in 4% paraformaldehyde and processed for immunohistochemical staining. For RNA and protein analysis, term villus tissues from the middle of the placenta were cut on the maternal side and a piece of tissue about 1 × 1 × 1 cm in size was spared, avoiding parts with blood clots, infarction, or calcification, to prepare both RNA and protein lysates. The patient characteristics are summarized in Table S1. All samples were randomly coded prior to processing to ensure that the analysts were blinded to their origin.
Cell culture
HTR-8/SVneo, an immortalized human trophoblast cell line derived from first-trimester human cytotrophoblast cells, was used for the study. HTR-8/SVneo cells were cultured in RPMI 1640 medium (C11875500BT, Gibco) supplemented with 10% fetal bovine serum (10099–141, Gibco) at 37 °C in a humidified incubator with 5% CO2 (Thermo Fisher Scientific).
Cell proliferation, invasion, and migration assays
Each group of cells was seeded into 96-well plates at a density of 5000 cells/well, and at 0, 24, 48, 72, and 96 h after seeding; four wells of each group of cells were tested using the Cell Counting Kit-8 (CK04, Dojindo Molecular Technologies, Kumamoto, Japan). The cell proliferation curve was plotted using Prism software. Cell invasion assays were carried out as described previously (8).
Quantitative real-time PCR (qPCR)
Total RNA was extracted from tissues using TRIzol (Invitrogen) according to the manufacturer's instructions, and cDNA was synthesized from 500 ng total RNA using a reverse transcription kit (Takara, Dalian, China). Quantitative real-time PCR was performed using an Applied Biosystems 7500 Sequence Detection System with SYBR Green chemistry, and GAPDH was used as an endogenous control. Each sample was used three times at different locations in the 96-well plate. Quantitative gene expression data were acquired and analyzed using the ABI Prism7500HT Sequence Detection System (RQ Manager, Grand Island, NY). All primers used in this study are listed in Table S2.
Western blot analysis
Tissue lysates were prepared using a radioimmunoprecipitation assay (RIPA) buffer. Equal amounts of protein (30 μg) were resolved by SDS-8/12% PAGE and electrotransferred to PVDF membranes. After blocking in TBS with Tween 20 (TBST) with 5% BSA for 2 h at room temperature, the membranes were incubated overnight with specific primary antibodies at 4 °C; the membranes were incubated with a primary TET2 antibody at a dilution of 1:500 (21207–1-AP, Proteintech Group), MMP9 antibody at a dilution of 1:200 (sc-393859, Santa Cruz Biotechnology), and normalized to β-tubulin as a control at dilution of 1:5000 (200608, Zen Bioscience). Primary antibody binding was visualized using horseradish peroxidase–conjugated goat anti-rabbit or anti-mouse IgG (1:10000, ZSGB-BIO). Signal intensities were measured using luminescent substrate for HRP (1305702, EMD Millipore Corporation) and Image J analysis software.
ELISA
Human MMP9 ELISA kit (EHC115.48, NeoBioscience) was used to determine MMP9 levels in 2 × 105 control, and TET2 knockdown HTR-8/SVneo cells were plated in the wells of a six-well plate with 4 ml DMEM without FBS. After 48 h, the conditioned medium was collected after centrifugation and used for the ELISA array analysis following the manufacturer's instructions. The absorbance was read using a microplate reader at a wavelength of 450 nm, and GraphPad Prism software was used for data analysis.
Dot-blot analysis
Genomic DNA was extracted from tissues using AxyPrep Multisource Genomic DNA miniprep kits (Axygen, Corning). A NanoDrop 2000 (Thermo Fisher Scientific) was used to quantify the DNA concentration. DNA samples (200 ng per sample) were diluted with 2 m NaOH and 10 mm Tris-Cl, pH 8.5; the DNA sample was serially diluted 2-fold and loaded on a nitrocellulose membrane (GE Healthcare). The blotted membrane was dried at 80 °C for 5 min and UV cross-linked at 120,000 μJ/cm2. The membrane was blocked in TBST with 5% BSA for 2 h at room temperature and then incubated overnight with mouse anti-5mC mAb (1:500, Active Motif) or mouse anti-5hmC mAb (1:5000, Abcam) at 4 °C. After washing, the membrane was incubated with HRP-conjugated anti-mouse/rabbit IgG (1:10000, ZSGB-BIO) secondary antibodies in TBST for 2 h at room temperature and signal intensities were measured using luminescent substrate for HRP (1305702, EMD Millipore) and Image J analysis software.
Human angiogenesis antibody array
The human angiogenesis antibody array kit was purchased from R&D Systems (ARY007). In total, 2 × 105 control and TET knockdown HTR-8/SVneo cells were plated in the wells of a six-well plate with 4 ml DMEM without FBS. After 48 h, the conditioned medium was collected after centrifugation and used for the human angiogenesis antibody array analysis following the manufacturer's instructions. In brief, the membranes were placed in a four-well multi-dish containing 2 ml of array blocking buffer. The membrane was incubated for 1 h on a rocking platform. Meanwhile, samples were mixed with the reconstituted Detection Antibody Mixture and incubated at room temperature for 1 h. After incubation, 1.5 ml of the sample/antibody mixture was added to the well. The membrane was incubated overnight at 4 °C on a rocking platform shaker, and then washed thrice using array wash buffer, with each wash lasting 10 min. Then, 2 ml of streptavidin-horseradish peroxidase solution was added onto the membrane and incubated for 30 min at room temperature on a rocking platform shaker. Next, three washes were performed with the array wash buffer. An ECL chemiluminescent substrate supplied with the kit was used to generate the signals and detailed pictures of the array were taken. Image J software was used to quantify the arrays.
Bisulfite sequencing analysis and pyrosequencing
Bisulfite sequencing analysis was performed as described previously (14). For sequence analysis, the BSP products were purified and subjected to direct sequencing by Tsingke Biotech (Chengdu, China). The CpG-rich region of the MMP9 promoter between −225 and −160 bp includes three CpG sites. The primer sequences used for pyrosequencing PCR are listed in Table S2.
ChIP
Magna ChIPTM G (17–611, EMD Millipore) was used to determine whether TET2 bound to MMP9 promoter in 2 × 107 HTR-8/SVneo cell. ChIP analysis followed the manufacturer's instructions. Protein-DNA complex was immunoprecipitated with TET2 antibody. IgG was used as an antibody control. The DNA was resuspended in water, and enzyme binding at MMP9 promoter was quantified by real-time qPCR using primers specific for MMP9 promoter. The target values were normalized to input controls.
Plasmid construction
The promoter sequence of MMP9 from −761 bp to the transcription start site shows the strongest transcriptional activity. The MMP9 promoter (780 bp) was amplified by PCR using Fast Pfu DNA polymerase (TransGen Biotech, Beijing, China) from HTR-8/SVneo cDNA before cloning into the pGL3-Basic vector. The MMP9 promoter was mutated from GCGC to TAAT using the Fast Mutagenesis System (TransGen Biotech). We constructed various plasmids based on WT of MMP9 promoter in the pGL3-Basic vector: all CpG sites methylated except −712 bp, which was demethylated by mutation (−712mut); all CpG sites methylated except −233 bp, which was demethylated by mutation (−233mut); and all CpG sites methylated except −712 bp and −233 bp site were demethylated by mutation (−712 and −233mut) (Fig. 3E). The plasmid of Tet2-CD and Tet2-HXD was generously donated by Prof. Chang (24). All the primers used in this study are listed in Table S2.
Dual-Luciferase reporter assays
The different DNA methylation statuses of pGL3-MMP9 vectors were determined by co-transfection with Renilla Luciferase vector into human embryo kidney 293T cells followed by measurement of luciferase activity using the Dual-Luciferase® Reporter Assay System (Promega).
Statistical analysis
All data were expressed as the mean ± S.D. All statistical analyses were performed using the GraphPad Prism software (version 5.00). A two-tailed Student's t test was used to compare two groups. Statistical comparisons were performed by one-way analysis of variance (ANOVA) for multiple groups. Significant differences between or among groups were indicated by *, p < 0.05 and **, p < 0.01.
Author contributions
X. Li, C. W., and W. X. conceptualization; X. Li, Y. S., L. T., and M. Z. formal analysis; X. Li, C. W., and W. X. writing-original draft; X. Li, C. W., and W. X. writing-review and editing; Y. S., M. Y., and T. P. data curation; K. W., L. T., T. P., X. Liu, and W. X. methodology; X. Liu investigation; W. X. supervision; W. X. project administration.
Supplementary Material
Acknowledgments
We thank all the women who participated in this study.
This work was supported by Natural Science Foundation of China Grants 81300494 and 81471463. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1 and S2.
- PE
- preeclampsia
- MMPs
- matrix metalloproteinases
- TET
- ten-eleven translocation enzymes
- BSP
- bisulfite sequencing PCR
- VC
- vitamin C
- TBST
- TBS with Tween 20.
References
- 1. American College of Obstetricians and Gynecologists; Task Force on Hypertension in Pregnancy (2013) Hypertension in pregnancy. Report of the American College of Obstetricians and Gynecologists' Task Force on Hypertension in Pregnancy. Obstet. Gynecol. 122, 1122–1131 10.1097/01.AOG.0000437382.03963.88 [DOI] [PubMed] [Google Scholar]
- 2. Abad C., Proverbio T., Piñero S., Botana D., Chiarello D. I., Marín R., and Proverbio F. (2012) Preeclampsia, placenta, oxidative stress, and PMCA. Hypertens. Pregnancy 31, 427–441 10.3109/10641955.2012.690058 [DOI] [PubMed] [Google Scholar]
- 3. Chaiworapongsa T., Chaemsaithong P., Korzeniewski S. J., Yeo L., and Romero R. (2014) Pre-eclampsia part 2: Prediction, prevention and management. Nat. Rev. Nephrol. 10, 531–540 10.1038/nrneph.2014.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. GBD 2015 Mortality and Causes of Death Collaborators. (2016) Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet 388, 1459–1544 10.1016/S0140-6736(16)31012-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Anderson C. M., Ralph J. L., Wright M. L., Linggi B., and Ohm J. E. (2014) DNA methylation as a biomarker for preeclampsia. Biol. Res. Nurs. 16, 409–420 10.1177/1099800413508645 [DOI] [PubMed] [Google Scholar]
- 6. Tang L., He G., Liu X., and Xu W. (2017) Progress in the understanding of the etiology and predictability of fetal growth restriction. Reproduction 153, R227–R240 10.1530/REP-16-0287 [DOI] [PubMed] [Google Scholar]
- 7. Lorquet S., Pequeux C., Munaut C., and Foidart J. M. (2010) Aetiology and physiopathology of preeclampsia and related forms. Acta Clin. Belg. 65, 237–241 10.1179/acb.2010.051 [DOI] [PubMed] [Google Scholar]
- 8. Yang M., Chen Y., Chen L., Wang K., Pan T., Liu X., and Xu W. (2016) miR-15b-AGO2 play a critical role in HTR8/SVneo invasion and in a model of angiogenesis defects related to inflammation. Placenta 41, 62–73 10.1016/j.placenta.2016.03.007 [DOI] [PubMed] [Google Scholar]
- 9. Cudmore M., Ahmad S., Al-Ani B., Fujisawa T., Coxall H., Chudasama K., Devey L. R., Wigmore S. J., Abbas A., Hewett P. W., and Ahmed A. (2007) Negative regulation of soluble Flt-1 and soluble endoglin release by heme oxygenase-1. Circulation 115, 1789–1797 10.1161/CIRCULATIONAHA.106.660134 [DOI] [PubMed] [Google Scholar]
- 10. Tanaka S., Nakanishi M. O., and Shiota K. (2014) DNA methylation and its role in the trophoblast cell lineage. Int. J. Dev. Biol. 58, 231–238 10.1387/ijdb.140053st [DOI] [PubMed] [Google Scholar]
- 11. Logan P. C., Mitchell M. D., and Lobie P. E. (2013) DNA methyltransferases and TETs in the regulation of differentiation and invasiveness of extra-villous trophoblasts. Front. Genet. 4, 265 10.3389/fgene.2013.00265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rahat B., Thakur S., Hamid A., Bagga R., and Kaur J. (2016) Association of aberrant methylation at promoter regions of tumor suppressor genes with placental pathologies. Epigenomics 8, 767–787 10.2217/epi.16.7 [DOI] [PubMed] [Google Scholar]
- 13. Rasmussen K. D., and Helin K. (2016) Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 30, 733–750 10.1101/gad.276568.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu H., Tang Y., Liu X., Zhou Q., Xiao X., Lan F., Li X., Hu R., Xiong Y., and Peng T. (2014) 14–3-3 tau (YWHAQ) gene promoter hypermethylation in human placenta of preeclampsia. Placenta 35, 981–988 10.1016/j.placenta.2014.09.016 [DOI] [PubMed] [Google Scholar]
- 15. Bowman R. L., and Levine R. L. (2017) TET2 in normal and malignant hematopoiesis. Cold Spring Harb. Perspect. Med. 7, a026518 10.1101/cshperspect.a026518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. An J., Rao A., and Ko M. (2017) TET family dioxygenases and DNA demethylation in stem cells and cancers. Exp. Mol. Med. 49, e323 10.1038/emm.2017.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Martin E., Ray P. D., Smeester L., Grace M. R., Boggess K., and Fry R. C. (2015) Epigenetics and preeclampsia: Defining functional epimutations in the preeclamptic placenta related to the TGF-β pathway. PLoS One 10, e0141294 10.1371/journal.pone.0141294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Palei A. C. T., Granger J. P., and Tanus-Santos J. E. (2013) Matrix metalloproteinases as drug targets in preeclampsia. Curr. Drug Targets 14, 325–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang Y., He G., Xu W., and Liu X. (2015) ENaC mediates human extravillous trophoblast cell line (HTR8/SVneo) invasion by regulating levels of matrix metalloproteinase 2 (MMP2). Placenta 36, 587–593 10.1016/j.placenta.2015.01.201 [DOI] [PubMed] [Google Scholar]
- 20. Plaks V., Rinkenberger J., Dai J., Flannery M., Sund M., Kanasaki K., Ni W., Kalluri R., and Werb Z. (2013) Matrix metalloproteinase-9 deficiency phenocopies features of preeclampsia and intrauterine growth restriction. Proc. Natl. Acad. Sci. U.S.A. 110, 11109–11114 10.1073/pnas.1309561110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fan M., Xu Y., Hong F., Gao X., Xin G., Hong H., Dong L., and Zhao X. (2016) Rac1/β-catenin signalling pathway contributes to trophoblast cell invasion by targeting Snail and MMP9. Cell Physiol. Biochem. 38, 1319–1332 10.1159/000443076 [DOI] [PubMed] [Google Scholar]
- 22. Peng B., Zhu H., Klausen C., Ma L., Wang Y. L., and Leung P. C. (2016) GnRH regulates trophoblast invasion via RUNX2-mediated MMP2/9 expression. Mol. Hum. Reprod. 22, 119–129 10.1093/molehr/gav070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ling L., Ren M., Yang C., Lao G., Chen L., Luo H., Feng Z., and Yan L. (2013) Role of site-specific DNA demethylation in TNFα-induced MMP9 expression in keratinocytes. J. Mol. Endocrinol. 50, 279–290 10.1530/JME-12-0172 [DOI] [PubMed] [Google Scholar]
- 24. Lio C. W., Zhang J., Gonzalez-Avalos E., Hogan P. G., Chang X., and Rao A. (2016) Tet2 and Tet3 cooperate with B-lineage transcription factors to regulate DNA modification and chromatin accessibility. eLife 5, e18290 10.7554/eLife.18290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pushpakumar S., Kundu S., Narayanan N., and Sen U. (2015) DNA hypermethylation in hyperhomocysteinemia contributes to abnormal extracellular matrix metabolism in the kidney. FASEB J. 29, 4713–4725 10.1096/fj.15-272443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tan Q., Wang W., Yang C., Zhang J., Sun K., Luo H. C., Mai L. F., Lao Y., Yan L., and Ren M. (2016) α-Ketoglutarate is associated with delayed wound healing in diabetes. Clin. Endocrinol. 85, 54–61 10.1111/cen.13047 [DOI] [PubMed] [Google Scholar]
- 27. Zhang J., Yang C., Wang C., Liu D., Lao G., Liang Y., Sun K., Luo H., Tan Q., Ren M., and Yan L. (2016) AGE-induced keratinocyte MMP-9 expression is linked to TET2-mediated CpG demethylation. Wound Repair Regen. 24, 489–500 10.1111/wrr.12426 [DOI] [PubMed] [Google Scholar]
- 28. Zhang Y. W., Wang Z., Xie W., Cai Y., Xia L., Easwaran H., Luo J., Yen R. C., Li Y., and Baylin S. B. (2017) Acetylation enhances TET2 function in protecting against abnormal DNA methylation during oxidative stress. Mol. Cell 65, 323–335 10.1016/j.molcel.2016.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kowluru R. A., Shan Y., and Mishra M. (2016) Dynamic DNA methylation of matrix metalloproteinase-9 in the development of diabetic retinopathy. Lab. Invest. 96, 1040–1049 10.1038/labinvest.2016.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Duraisamy A. J., Mishra M., and Kowluru R. A. (2017) Crosstalk between histone and DNA methylation in regulation of retinal matrix metalloproteinase-9 in diabetes. Invest. Ophthalmol. Vis. Sci. 58, 6440–6448 10.1167/iovs.17-22706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nelissen E. C., van Montfoort A. P., Dumoulin J. C., and Evers J. L. (2011) Epigenetics and the placenta. Hum. Reprod. Update 17, 397–417 10.1093/humupd/dmq052 [DOI] [PubMed] [Google Scholar]
- 32. Gao W. L., Li D., Xiao Z. X., Liao Q. P., Yang H. X., Li Y. X., Ji L., and Wang Y. L. (2011) Detection of global DNA methylation and paternally imprinted H19 gene methylation in preeclamptic placentas. Hypertens. Res. 34, 655–661 10.1038/hr.2011.9 [DOI] [PubMed] [Google Scholar]
- 33. Takayama K., Misawa A., Suzuki T., Takagi K., Hayashizaki Y., Fujimura T., Homma Y., Takahashi S., Urano T., and Inoue S. (2015) TET2 repression by androgen hormone regulates global hydroxymethylation status and prostate cancer progression. Nat. Commun. 6, 8219 10.1038/ncomms9219 [DOI] [PubMed] [Google Scholar]
- 34. Chinnathambi V., Balakrishnan M., Ramadoss J., Yallampalli C., and Sathishkumar K. (2013) Testosterone alters maternal vascular adaptations: role of the endothelial NO system. Hypertension 61, 647–654 10.1161/HYPERTENSIONAHA.111.00486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Qu K., Ma X. F., Li G. H., Zhang H., Liu Y. M., Zhang K., Zeng J. F., Lei J. J., Wei D. H., and Wang Z. (2017) Vitamin C down-regulate apo(a) expression via Tet2-dependent DNA demethylation in HepG2 cells. Int. J. Biol. Macromol. 98, 637–645 10.1016/j.ijbiomac.2017.02.025 [DOI] [PubMed] [Google Scholar]
- 36. Agathocleous M., Meacham C. E., Burgess R. J., Piskounova E., Zhao Z., Crane G. M., Cowin B. L., Bruner E., Murphy M. M., Chen W., Spangrude G. J., Hu Z., DeBerardinis R. J., and Morrison S. J. (2017) Ascorbate regulates haematopoietic stem cell function and leukaemogenesis. Nature 549, 476–481 10.1038/nature23876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cimmino L., Dolgalev I., Wang Y., Yoshimi A., Martin G. H., Wang J., Ng V., Xia B., Witkowski M. T., Mitchell-Flack M., Grillo I., Bakogianni S., Ndiaye-Lobry D., Martín M. T., Guillamot M., et al. (2017) Restoration of TET2 function blocks aberrant self-renewal and leukemia progression. Cell 170, 1079–1095.e20 10.1016/j.cell.2017.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Azami M., Azadi T., Farhang S., Rahmati S., and Pourtaghi K. (2017) The effects of multi mineral-vitamin D and vitamins (C+E) supplementation in the prevention of preeclampsia: An RCT. Int. J. Reprod. Biomed. 15, 273–278 [PMC free article] [PubMed] [Google Scholar]
- 39. Shorey-Kendrick L. E., McEvoy C. T., Ferguson B., Burchard J., Park B. S., Gao L., Vuylsteke B. H., Milner K. F., Morris C. D., and Spindel E. R. (2017) Vitamin C prevents offspring DNA methylation changes associated with maternal smoking in pregnancy. Am. J. Respir. Crit. Care Med. 196, 745–755 10.1164/rccm.201610-2141OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhu L., Lv R., Kong L., Cheng H., Lan F., and Li X. (2015) Genome-wide mapping of 5mC and 5hmC identified differentially modified genomic regions in late-onset severe preeclampsia: A pilot study. PLoS One 10, e0134119 10.1371/journal.pone.0134119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. White W. M., Brost B., Sun Z., Rose C., Craici I., Wagner S. J., Turner S. T., and Garovic V. D. (2013) Genome-wide methylation profiling demonstrates hypermethylation in maternal leukocyte DNA in preeclamptic compared with normotensive pregnancies. Hypertens. Pregnancy 32, 257–269 10.3109/10641955.2013.796970 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.