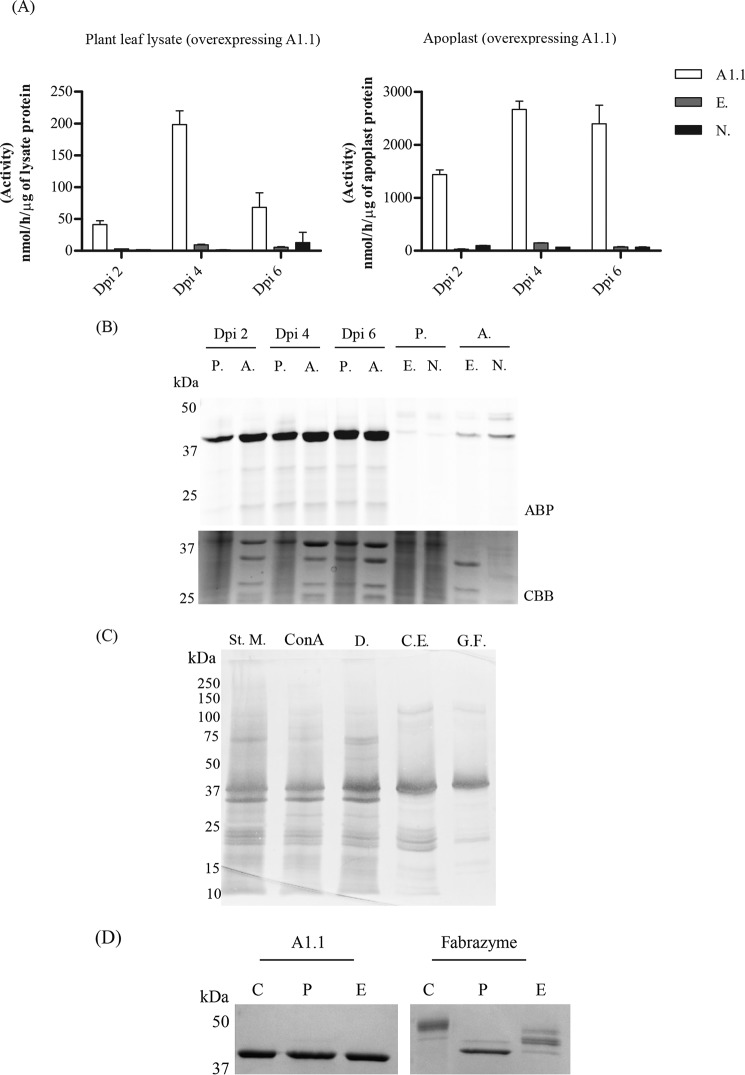
Figure 3.
Overexpression and biochemical characterization of the newly identified N. benthamiana α-galactosidase, A1.1. A, transient overexpression of A1.1 in N. benthamiana leaves via A. tumefaciens transformation assays. Infiltrated leaves and apoplast samples were harvested on different days post-infiltration (dpi): 2, 4, and 6. The expression levels of the enzyme were first tested via 4MU–α-Gal activities in lysates and apoplast fractions of different dpi. n = 2. (A1.1 = leaf overexpressing A1.1; E. = empty vector; N. = nontreated plant leaf.) B, next, the expression levels of active enzyme molecules were tested via in-gel ABP labeling. CBB staining of the gels followed to ensure the presence of the overexpressed enzyme. (P. = plant lysate; A. = apoplast sample; E. = empty vector; N. = nontreated plant leaf.) C, purification overview. SDS-PAGE analysis following silver staining of fractions obtained during different purification steps. (St. M. = starting material; ConA = fraction not bound to concanavalin A column; D. = sample obtained after 48 h of dialysis; C.E. = pooled collected eluate from cation-exchange chromatography; G.F. = final pooled fraction obtained after gel filtration.) 2 μg of total protein were loaded in each lane, except in G.F., where 1 μg was loaded. D, SDS-PAGE analysis following Coomassie Brilliant Blue staining of 1 μg of pure A1.1 and Fabrazyme after treatment with PNGase F and EndoH (C = untreated pure enzyme; P = enzyme treated with PNGase F; E = enzyme treated with EndoH), shows that A1.1 is not likely N-glycosylated, whereas Fabrazyme is, due to the difference in molecular weight after treatment.