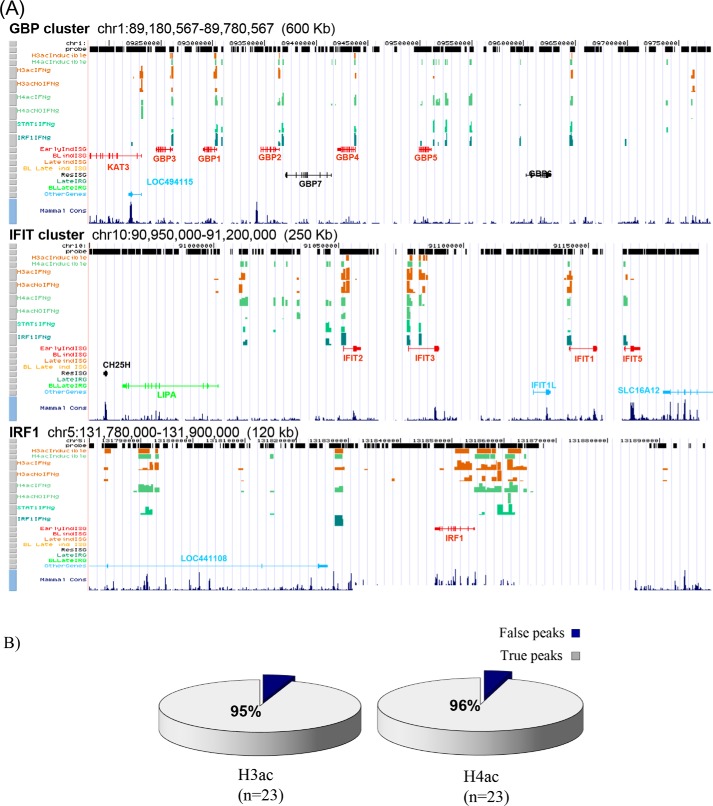
Figure 1.
Mapping constitutive and IFNγ-induced chromatin acetylation at IFNγ-induced STAT1- and IRF1-binding sites. A, chromatin acetylation maps. Tracks are organized in the following order from the top: chromosomal coordinates, probe positions, inducible histone acetylation (H3ac- and H4ac-inducible), plus and minus IFNγ tracks for H3ac (H3acIFNg and H3acNoIFNg) and H4ac (H4acIFNg and H4acNoIFNg), and only the plus IFNg track for STAT1 (STAT1IFNg) and IRF1 (IRF1IFNg) (from Ref. 11). Gene tracks are categorized according to their responses to IFNγ (from Ref. 11) and presented in different colors. Early, 6 h; Late, 24 h; ind, induced; BL, borderline; Res, resistant. Mammal Cons track shows the degree of sequence conservation between 44 vertebrate species. Additional acetylation maps are provided in Fig. S1. B, ChIP–qPCR validation. Arbitrarily selected ChIP-chip H3ac and H4ac sites were examined by ChIP–qPCR on chromatin from HeLa cells with no or 6 h of IFN treatment. ≥95% of peaks were validated in both cases.