Preterm birth is associated with both psychological and physical disabilities and is the leading cause of infant morbidity and mortality worldwide. Infection is known to be an important cause of spontaneous preterm birth, and recent research has implicated variation in the “placental microbiome” in the risk of preterm birth. Consistent with data from previous studies, the abundances of certain clinically relevant species differed between spontaneous preterm- and nonspontaneous preterm- or term-delivered placentas. These results support the view that a proportion of spontaneous preterm births have an intrauterine-infection component. However, an additional observation from this study was that a substantial proportion of sequenced reads were contaminating reads rather than DNA from endogenous, clinically relevant species. This observation warrants caution in the interpretation of sequencing outputs from low-biomass samples such as the placenta.
KEYWORDS: contamination, infection, microbiome, pregnancy, preterm birth
ABSTRACT
In this study, differences in the placental microbiota from term and preterm deliveries in a large pregnancy cohort in the United Kingdom were studied by using 16S-targeted amplicon sequencing. The impacts of contamination from DNA extraction, PCR reagents, and the delivery itself were also examined. A total of 400 placental samples from 256 singleton pregnancies were analyzed, and differences between spontaneous preterm-, nonspontaneous preterm-, and term-delivered placentas were investigated. DNA from recently delivered placentas was extracted, and screening for bacterial DNA was carried out by using targeted sequencing of the 16S rRNA gene on the Illumina MiSeq platform. Sequenced reads were analyzed for the presence of contaminating operational taxonomic units (OTUs) identified via sequencing of negative extraction and PCR-blank samples. Differential abundances and between-sample (beta) diversity metrics were then compared. A large proportion of the reads sequenced from the extracted placental samples mapped to OTUs that were also found for negative extractions. Striking differences in the compositions of samples were also observed, according to whether the placenta was delivered abdominally or vaginally, providing strong circumstantial evidence for delivery contamination as an important contributor to observed microbial profiles. When OTU- and genus-level abundances were compared between the groups of interest, a number of organisms were enriched in the spontaneous preterm-delivery cohort, including organisms that have been associated previously with adverse pregnancy outcomes, specifically Mycoplasma spp. and Ureaplasma spp. However, analyses of the overall community structure did not reveal convincing evidence for the existence of a reproducible “preterm placental microbiome.”
IMPORTANCE Preterm birth is associated with both psychological and physical disabilities and is the leading cause of infant morbidity and mortality worldwide. Infection is known to be an important cause of spontaneous preterm birth, and recent research has implicated variation in the “placental microbiome” in the risk of preterm birth. Consistent with data from previous studies, the abundances of certain clinically relevant species differed between spontaneous preterm- and nonspontaneous preterm- or term-delivered placentas. These results support the view that a proportion of spontaneous preterm births have an intrauterine-infection component. However, an additional observation from this study was that a substantial proportion of sequenced reads were contaminating reads rather than DNA from endogenous, clinically relevant species. This observation warrants caution in the interpretation of sequencing outputs from low-biomass samples such as the placenta.
INTRODUCTION
Preterm birth (PTB), defined as any delivery before 37 completed weeks of gestation, affects between 5 and 18% of pregnancies. It is the leading cause of neonatal morbidity and mortality worldwide and inflicts substantial physical, psychological, and economic costs upon affected families and wider society. Epidemiological studies implicate factors such as maternal ethnicity (1–3), age (4), body mass index (BMI) (5–7), and smoking (8). However, a detailed etiological understanding of spontaneous PTB (sPTB) remains limited.
An association between bacterial infection and sPTB has long been hypothesized, and evidence for its involvement continues to grow. It is estimated that between 25% and 40% of sPTBs involve intrauterine infection (8). This proportion increases steadily as the gestational age (GA) at birth decreases and may be a mediator in as many as 79% of births at 23 weeks of gestation (9). Infection is hypothesized to lead to PTB by eliciting an inflammatory response in the mother and/or fetus, triggering early labor and/or membrane rupture (10).
Studies using both culture-based and molecular techniques have reported associations between sPTB and the presence or profile of bacteria in amniotic fluid (11–13), the placental parenchyma (14–16), fetal membranes (16–18), the placental basal plate (19), cord blood (13), cervical fluid (20), and maternal vaginal flora (21). The majority of organisms recovered, such as Mycoplasma hominis, implicate routes of infection that originate in the vagina. However, the identification of commensal oral species in intrauterine tissues, notably Fusobacterium nucleatum, also implicates maternal oral health and the hematogenous spread of organisms to the uterine cavity in the risk of sPTB (13, 18, 22).
The majority of studies reporting the presence of bacteria in intrauterine tissues have been designed to investigate the etiology of adverse pregnancy outcomes. However, the recovery of bacteria in healthy pregnancies has also been reported (14, 18, 19, 23). Some authors have interpreted those observations as evidence that a “maternal microbiome” may be a functional component of normal human pregnancy (14, 24). This has sparked considerable discussion regarding the meaning, reliability, and frequency of such nonpathogenic colonization (24–29). It is a technical and statistical challenge to reliably differentiate endogenous, clinically meaningful bacterial DNA from contamination picked up during delivery or sample collection/preparation (30, 31). Indeed, a recent study observed no difference in the microbial signatures from contamination in controls obtained at all stages of collection and sampling and DNA extracted from placental samples (32).
We conducted a nested case-control study of placental samples from term and preterm deliveries to explore the nature of intrauterine bacterial colonization in healthy and adverse pregnancies. It was hypothesized that placental samples taken from pregnancies culminating in sPTB would harbor microbial profiles distinct from those from placental samples taken from pregnancies culminating in nonspontaneous preterm birth (nsPTB) or term birth. Using samples from a large, United Kingdom-based pregnancy cohort, the Baby Bio Bank (BBB), the composition and structure of bacterial communities recovered from samples from preterm and term births were assessed in order to test this hypothesis, using targeted 16S amplicon sequencing. The impact of reagent contamination on the sequencing data set was also investigated.
RESULTS
Subject demographics and sampling types.
A total of 400 samples from 256 pregnancies were sequenced for this study, 50 of which were nonspontaneous preterm deliveries, 41 of which were spontaneous preterm deliveries, and 165 of which were term deliveries. The majority (89%) of biological samples were obtained from parenchyma tissue, and a small number of pregnancies also had matching villous tissue, from the maternal side of the placenta, available for sequencing. A summary of key maternal characteristics for the whole cohort is outlined in Table 1.
TABLE 1.
Selection of demographic and clinical characteristics of the sequenced cohort
| Maternal characteristic | Value for group |
||||
|---|---|---|---|---|---|
| sPTB (n = 41) | nsPTB (n = 50) | Term (n = 165) | All (n = 256) | Missing | |
| Median GA at birth (wk) (range) | 35 (23–36) | 35 (26–36) | 40 (37–42) | 40 (23–42) | |
| No. (%) of vaginal deliveries | 31 (75.6) | 5 (10) | 84 (63.2) | 120 (53.6) | 32 |
| No. (%) of individuals of maternal white ethnicity | 27 (65.9) | 25 (50.0) | 131 (79.4) | 183 (71.5) | |
| No. (%) of nonsmokers | 39 (95.1) | 45 (90.0) | 157 (95.2) | 241 (94.1) | |
| No. (%) of individuals with maternal obesity | 6 (14.6) | 10 (20) | 29 (17.9) | 45 (17.8) | 3 |
Significant proportions of total reads are reagent contaminants.
We attempted to differentiate sequences amplified from original, endogenous bacterial DNA present in placental samples at delivery from contaminating reads. A total of 136 operational taxonomic units (OTUs) from 44 genera (see Table S1 in the supplemental material for a full list of contaminating genera identified) were flagged by using our definition of potential contaminants (see Materials and Methods). A total of 32 (73%) of these genera were reported previously to be reagent contaminants (30).
Some flagged contaminants present in negative extractions may have originated from the experimental samples themselves, rather than extraction kits. Sample-to-control crossover due to false index pairings during PCR was reported previously (33, 34). On the basis of previous evidence and comparisons of relative abundances between negative and experimental samples, potential contaminant OTUs mapping to Lactobacillus spp., Veillonella spp., and Mycoplasma spp. were considered erroneously flagged and were not removed from samples.
The remaining “potentially contaminating” OTUs were removed from downstream analyses. This approach led to a substantial reduction in the size of the sample data set, discarding 933,083 reads, 20.3% of the total, and 132 OTUs from experimental samples. Following error checking and contaminant filtering, any sample with <500 reads was removed from the data set, and technical replicates were merged. A total of 3,590,138 reads were retained, mapping to 261 unique biological samples from 199 pregnancies (40 nsPTB, 33 sPTB, and 126 term samples). Prior to filtering, the median number of reads per sample was 2,831 (interquartile range [IQR] = 976.5 to 8,741), and following filtering, this value was 2,526 (IQR = 1,166 to 8,479). There was no difference in the distributions of subject groupings following the removal of contaminant OTUs (chi-squared value = 0.17; P = 0.92). Following filtering, 146 pregnancies retained 1 biological replicate, 44 pregnancies had 2, and 9 pregnancies had 3.
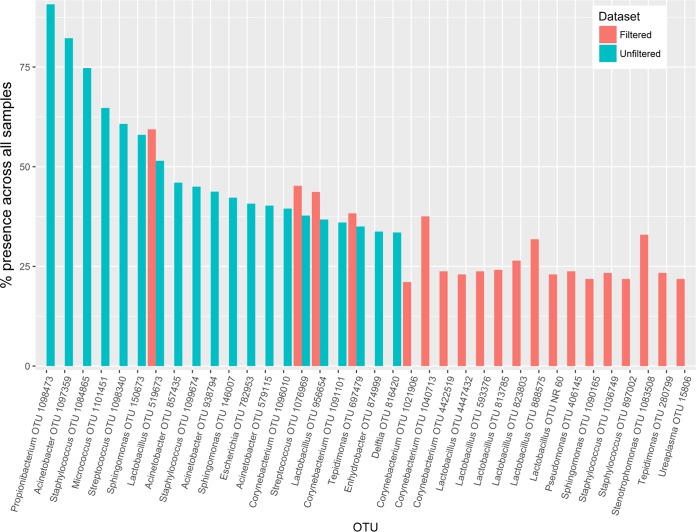
The removal of contaminant reads and low-abundance samples led to an observable shift in the taxonomic composition of the data set. This can be seen by comparing the rank abundance curves for the 20 most widely abundant OTUs before and after filtering, between which only 4 OTUs were shared (Fig. 1). The most widely abundant OTU in the nonfiltered sample set mapped to the skin commensal Propionibacterium acnes and was present in 90.8% of the samples. In contrast, the most widely abundant OTU in the filtered data set mapped to Lactobacillus crispatus and was present in only 59.4% of samples. Eight of the 20 top OTUs in the filtered data set mapped to the Lactobacillus genus.
FIG 1.
Rank abundances of top 20 OTUs in filtered and unfiltered data sets. The 20 most widely abundant OTUs in the data sets before and after filtering for negative contaminants were identified, and their percent presences across the cohort are compared. Only 4 OTUs were shared between the filtered and unfiltered data sets, and many OTUs that were removed mapped to known contaminants.
Delivery method is influential for certain highly abundant genera.
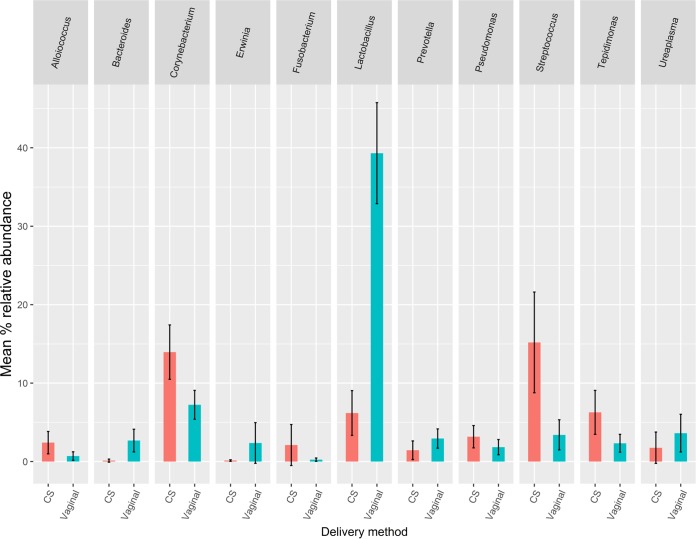
The 20 most widely abundant OTUs in the filtered data set were all present in >20% of samples (Fig. 1). However, when the taxonomic makeups of vaginally delivered and caesarean section (CS)-delivered placentas were compared, clear differences were observed. The seven genera with the highest mean relative abundances from CS deliveries combined with the seven genera with highest mean relative abundances from vaginal deliveries are shown in Fig. 2. A number of these highly abundant genera were shared between CS delivery and vaginal delivery groups. However, when the relative abundances of these genera were compared between the two groups, 6 were significantly differentially abundant by delivery method (Table 2). Two common vaginal genera, Lactobacillus and Bacteroides, were significantly more abundant in samples from vaginal deliveries. In contrast, the common skin flora genera Streptococcus and Corynebacterium were present at significantly higher abundance in CS samples.
FIG 2.
Top 11 genera from CS (red) and vaginal (blue) deliveries with the highest mean relative abundances. Genera were combined to form 11 unique groups, and the mean relative abundances between the two groups were compared and are shown. Error bars show 95% confidence intervals around the means.
TABLE 2.
Differences in mean relative abundances of the 11 CS and vaginal genera with the highest mean relative abundancesa
| Genus | Difference in % relative abundance (CS − vaginal) | P value |
|---|---|---|
| Lactobacillus | −33.12 | 1.04E−16* |
| Bacteroides | −2.55 | 8.17E−04* |
| Erwinia | −2.24 | 9.36E−02 |
| Ureaplasma | −1.86 | 2.45E−01 |
| Prevotella | −1.49 | 8.90E−02 |
| Pseudomonas | 1.33 | 1.31E−01 |
| Alloiococcus | 1.71 | 2.99E−02* |
| Fusobacterium | 1.86 | 1.68E−01 |
| Tepidimonas | 3.94 | 1.19E−02* |
| Corynebacterium | 6.72 | 1.03E−03* |
| Streptococcus | 11.79 | 8.09E−04* |
Student's t test and a two-sided test for significance were used for comparison of means (*, P < 0.05).
sPTB is associated with novel and established genera from placental tissue.
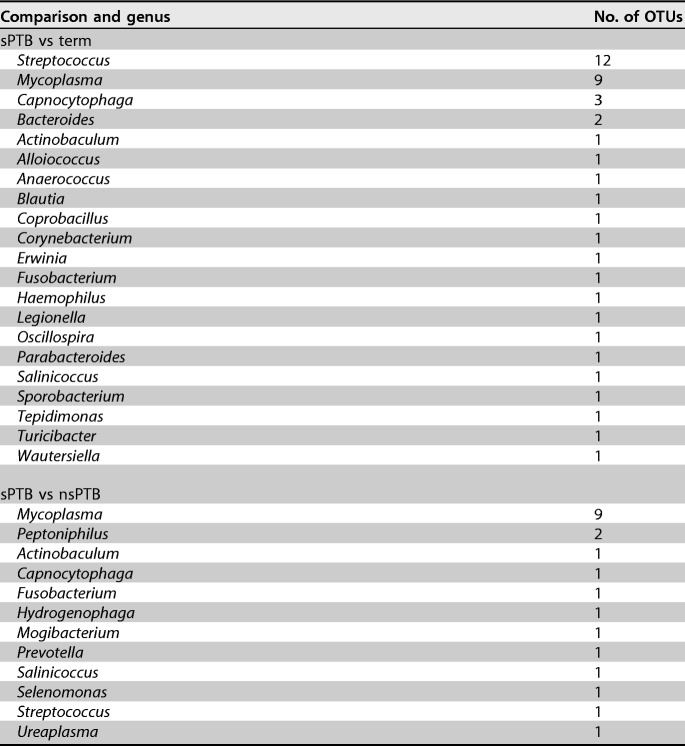
The primary hypothesis of this study was that certain organisms would be differentially abundant in placental tissue according to pregnancy outcome. Therefore, we compared the abundances of OTUs and genera in placental tissues from sPTB pregnancies with those from term or nsPTB pregnancies. Univariate analyses were first run on the 261 remaining filtered samples (41 sPTB, 47 nsPTB, and 173 term samples). Six genera had a significantly higher abundances (P < 0.01) in sPTB tissue than in nsPTB tissue (Ureaplasma, Prevotella, Salinicoccus, Mycoplasma, Capnocytophaga, and Anaerococcus), and seven genera were more highly abundant in sPTB than in term samples (Tepidimonas, Salinicoccus, Capnocytophaga, Mycoplasma, Anaerococcus, Truepera, and Coprobacillus) (see Tables S2 and S3 in the supplemental material for full results of unadjusted comparisons).
Models were then adjusted for the potential confounding effects of delivery method, recruiting hospital, maternal ethnicity, BMI, smoking behavior, and tissue type. These confounders were selected on the basis of previous evidence of associations with both gestation length and microbiome profiles. This adjusted cohort was smaller than that used for univariate analyses (total, n = 219; sPTB, n = 41; nsPTB, n = 47; term birth, n = 131) due to missing data on delivery method and maternal BMI.
The genera of those OTUs with high abundances in sPTB placentas, compared to either nsPTB or term deliveries following adjustment, and with P values of <0.01 are listed in Table 3 (full results are listed in Tables S4 and S5 in the supplemental material). When this comparison was repeated with all OTUs pooled at the genus level, 4 genera were found at significantly higher abundances in sPTB than in nsPTB placentas at a threshold P value of <0.01 (Mycoplasma, Ureaplasma, Mogibacterium, and Salinicoccus) (Table 4). Eight genera were more highly abundant in the sPTB-versus-term comparisons at the same threshold (Anaerococcus, Capnocytophaga, Coprobacillus, Erwinia, Mycoplasma, Salinicoccus, Turicibacter, and Tepidimonas) (Table 5).
TABLE 3.
Genera and numbers of individual OTUs enriched in sPTB placentas compared to term and nsPTB placentas following adjustment for potential confounders, with a P value of <0.01
TABLE 4.
Genera enriched in sPTB versus nsPTB placentas following adjustment, with a P value of <0.01
| Genus | Log2 fold change (95% CI)a | P | Q |
|---|---|---|---|
| Ureaplasma | 3.42 (1.86–4.99) | 2.58E−05 | 6.50E−03 |
| Mycoplasma | 1.84 (0.66–3.01) | 2.38E−03 | 2.00E−01 |
| Mogibacterium | 0.73 (0.23–1.22) | 4.22E−03 | 2.66E−01 |
| Salinicoccus | 0.48 (0.12–0.84) | 8.64E−03 | 3.11E−01 |
CI, confidence interval.
TABLE 5.
Genera enriched in sPTB versus term placentas following adjustment, with a P value of <0.01
| Genus | Log2 fold change (95% CI) | P | Q |
|---|---|---|---|
| Capnocytophaga | 1.03 (0.53–1.52) | 6.45E−05 | 1.62E−02 |
| Tepidimonas | 2.31 (0.97–3.65) | 8.04E−04 | 1.01E−01 |
| Salinicoccus | 0.46 (0.17–0.74) | 1.84E−03 | 1.16E−01 |
| Coprobacillus | 0.22 (0.07–0.36) | 3.39E−03 | 1.71E−01 |
| Mycoplasma | 1.36 (0.42–2.3) | 4.60E−03 | 1.93E−01 |
| Anaerococcus | 1.61 (0.46–2.76) | 6.27E−03 | 2.26E−01 |
| Turicibacter | 0.36 (0.1–0.62) | 7.94E−03 | 2.50E−01 |
| Erwinia | 1.23 (0.3–2.17) | 9.74E−03 | 2.65E−01 |
Beta diversity does not differentiate between pregnancy outcomes.
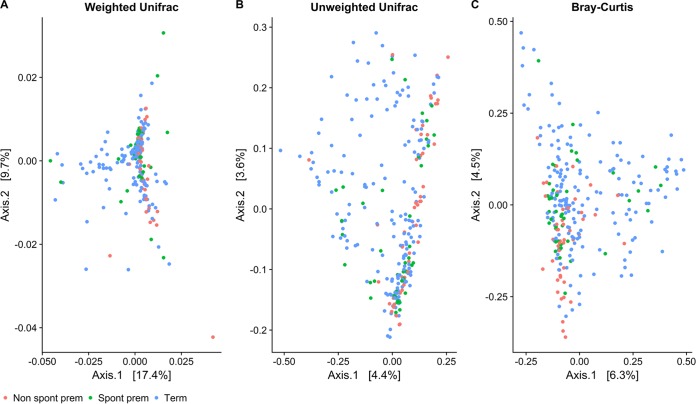
Recent work has suggested that a recognizable shift in structure of an overall “placental microbiome” may be observable in beta diversity comparisons between the different outcome groups (14). In order to investigate this hypothesis in our cohort, weighted UniFrac, unweighted UniFrac, and Bray-Curtis distance matrices were produced from the data to estimate beta diversity. Distances were then plotted by using principal-component analysis (PCoA) to visualize the first two axes and colored by pregnancy outcome (Fig. 3). Samples did not clearly cluster by pregnancy outcome with any of the methods used. Results from analyses conducted to quantify differences in beta diversity between outcomes produced very similar R2 values, all of which were significant at a P value of 0.001 (see Table S6 in the supplemental material). However, only a very small proportion of the variance between samples (∼2%) was accounted for by these groupings using this method.
FIG 3.
PCoA ordination of 3 beta diversity matrices. Shown are the first two PCoA axes that were plotted from weighted UniFrac (A), unweighted UniFrac (B), and Bray-Curtis (C) distances using VST-normalized counts of all samples in the cohort to compare beta diversity values between groups of interest.
DISCUSSION
The results from this study show that bacterial DNA from a variety of organisms was present in placental samples taken from a cohort of both normal and complicated pregnancies in the United Kingdom. This observation of a low-level, relatively diverse placental microbial signature is supported by data from other recent molecular studies working with the same tissues (14, 16, 18, 23, 35). A number of organisms of potential clinical relevance were also shown to be enriched in sPTB placentas compared to nsPTB and term tissues. However, we also observed widespread contamination across all placental samples, regardless of clinical outcome. These observations implicated both the delivery method and reagent contamination as significant contributors to our overall sequencing output. In addition, beta diversity analyses did not support the existence of a “unique preterm microbiome” in terms of a structured community shared among this particular obstetric group and distinguishable from other outcome groups. The very subtle differences observed in overall taxonomic compositions between clinical groups, in conjunction with the unclustered data in the PCoA plots, imply that such variation is unlikely to be clinically relevant.
Sequencing of a tissue with a low microbial biomass, such as the placenta, is a significant methodological and statistical challenge. In order to understand the clinical significance of those organisms identified by 16S sequencing, establishing the provenance of the sequenced DNA is critical. The presence of nonendogenous transcripts in sequencing outputs could have been the result of true bacterial contamination picked up during sample collection and experimental preparation processes or a function of PCR and sequencing artifacts. It was clear from our analyses that a significant proportion of the DNA identified was likely acquired during sample preparation, rather than being representative of true endogenous colonization. As a result, many samples were removed after filtering, as they did not yield sufficient noncontaminant DNA for analysis. Only four of the most widely abundant OTUs in the nonfiltered data set were also present in the filtered data set. Overall, the filtered OTUs were spread less widely across the sample sets, which is in line with a conceptualization of contaminants as organisms that are likely to affect all samples equally. These data demonstrate the need to account for contaminants in microbiome studies and the potentially erroneous conclusions that could be drawn from the data if their impact is ignored, as has been noted previously by other authors (29, 32).
Delivery method is a potentially important confounder in associations between GA at birth and placental microbial profiles. In our data, the highest proportion of vaginal deliveries was in the sPTB group, and the lowest proportion was in the nsPTB group. This trend may reflect the higher incidence of clinical indications, such as preeclampsia among nsPTB pregnancies, which require swift delivery to protect the health of the mother and baby. In our study, many OTUs were shared across placental samples, which could be interpreted as support for the existence of a common placental microbiome across pregnancies. However, it is notable that many of these shared OTUs mapped to skin and vaginal commensals that clearly varied by delivery method. Such comparisons in our study support the hypothesis that much of the signal observed in our data set may reflect contamination picked up during delivery (vaginal or CS), rather than a truly endogenous placental microbiome. These observations highlight the importance of accounting for delivery method in differential-abundance comparisons.
This study provided evidence for the enrichment of a number of organisms within sPTB placental samples, independent of the mode of delivery. Although the majority of OTU and genus groupings did not reach statistical significance once adjusted for multiple testing (Q > 0.1), some of the most significantly differentiated genera, such as Mycoplasma and Ureaplasma spp., were reported previously to be opportunistic intrauterine pathogens highly correlated with the incidence of sPTB (18, 36–41), supporting the output from our study. Others, such as Capnocytophaga, have been less well studied with respect to sPTB pathogenesis. Interestingly, the main organisms that were associated with PTB in the most widely cited placental microbiome study by Aagaard et al. (14), such as Burkholderia spp., did not overlap those found here.
Capnocytophaga was present across a number of samples at a relatively low total abundance compared to genera such as Mycoplasma. However, it was one of only two genera, along with the well-known PTB-associated organism Ureaplasma, that remained significantly associated with sPTB placentas following adjustment for multiple testing. This anaerobic organism is usually isolated from the oral cavity and is rarely isolated from the genital tract. However, it was previously associated with intrauterine infection in a few reports (42–44). Outside pregnancy, Capnocytophaga infections tend to be most commonly reported in immunocompromised children and are known to be involved in periodontitis (45), which is interesting given the immunosuppressed state of pregnancy.
Dissection and extraction of samples for this study were carried out by using rigorously controlled, sterile procedures. However, our placental samples were not originally collected with the intention to be used in microbial analyses. We acknowledge that this is a limitation of our study. Similarly, the storage and cleaning reagents RNAlater and phosphate-buffered saline (PBS), which were used by the collection team, were not available for use as comparative sequencing controls, as the extraction reagents were. Therefore, the estimation of contaminating reads carried out during the data cleanup stage may have been an underestimate of the true burden of exogenous bacterial reads in our placental data set. However, adjustment in final models for hospital collection site would account for at least some site-specific contamination patterns.
The specific study of the placental microbiome remains in its infancy. This research and other similar data suggest that there may be a low-level nonpathogenic placental microbiome present in many, if not all, placentas. However, differentiating this from organisms picked up at delivery or during experimental handling is an ongoing challenge. In addition, analyses of the overall community structure in our samples did not reveal convincing evidence for the existence of a reproducible “preterm placental microbiome.”
Our study provides one of the largest cohorts of 16S-sequenced placental tissue from sPTBs in the literature. This study gathered novel data on a tissue that remains relatively unexplored, from an unbiased microbiological perspective. The cohort consisted of a large number of spontaneous, early preterm births, providing a powerful opportunity to detect colonization patterns relevant to adverse pregnancy outcomes. Furthermore, the use of a specifically defined “nonspontaneous” preterm birth group was a novel addition. These nsPTB placentas provided a comparison group that was essentially matched for GA with the sPTB cases but very likely had a different underlying etiology. Further work is required to elucidate the clinical significance of specific organisms identified here for sPTB initiation and develop more-targeted strategies to mitigate their pathogenic effect.
MATERIALS AND METHODS
Study design and recruitment.
Samples used in this study were taken from the large United Kingdom-based resource for research into complications in pregnancy, the Baby Bio Bank (BBB) (https://www.ucl.ac.uk/tapb/sample-and-data-collections-at-ucl/biobanks-ucl/baby-biobank). Recruitment and sample collection occurred at Queen Charlotte and Chelsea, Chelsea and Westminster, and St Mary's hospitals in London. Detailed descriptions of the contents of the bank and sample collection procedures were reported in a previous study (46). Written informed consent was obtained from participants in advance of delivery. Preterm births were defined as any delivery at <37 weeks of gestation. Subjects were further divided into sPTB and nsPTB subcategories by using available clinical data on labor and delivery. sPTB was defined as any delivery precipitated by spontaneous labor and/or spontaneous membrane rupture. Nonspontaneous events consisted of artificial or no membrane rupture, combined with induced or no labor events, and were often precipitated by maternal clinical events such as preeclampsia.
Placental sample collection and storage.
All placental samples were collected by the hospital's maternity team and dissected by a BBB recruiter following delivery. For each placenta, 1-cm3 specimens were excised from four points below the membrane on the chorionic plate (placental parenchyma), close to the umbilical cord entrance. Villous tissue pooled from 6 sites on the maternal basal plate and combined into one collection tube was also collected from a subset of placentas. All samples were rinsed in PBS to remove excess maternal blood, placed into barcoded cryogenic tubes along with 5 ml of RNAlater, and stored at −80°C.
Placental DNA extraction.
A total of 20 to 50 mg of either villous or chorionic plate placental tissue was excised from stored samples in a sterile laminar flow tissue culture hood, using sterile disposable scalpels and petri dishes. Total DNA was then extracted by using the Qiagen DNeasy blood and tissue kit, according to the manufacturer's instructions, with an additional bead-beating step with MPBio Lysing Matrix B beads to minimize Gram-negative extraction bias (47, 48). All DNA was stored at −20°C until required. A negative extraction control, in which no tissue was added to extraction reagents and the normal protocol was carried out, was produced for every round of extractions.
16S rRNA gene high-throughput sequencing.
Libraries were prepared by using primers targeting the V5-V7 regions of the 16S rRNA gene (785F [5′-GGATTAGATACCCBRGTAGTC-3′] and 1175R [5′-ACGTCRTCCCCDCCTTCCTC-3′]) (16, 35). Primers were adapted for high-throughput sequencing with the addition of Illumina P5 or P7 adapter sequences and barcoded dual-index forward and reverse sequences taken from a previous study (49). Ultrapure Taq DNA polymerase (Molzym) was used to minimize the chance of contamination of the sequencing library from bacteria present in PCR reagents, and a 32-cycle endpoint PCR was run to amplify the bacterial template (see Tables 6 and 7 for reaction components and specific cycling conditions, respectively). PCR products were double cleaned by using 0.8× AMPure XP beads, according to the manufacturer's instructions. Custom primers were loaded onto an Illumina MiSeq instrument along with the cleaned and diluted library for a 500-cycle V2 kit.
TABLE 6.
Reaction components for preparation of the library for 16S amplicon sequencing of placental samplesa
| Reaction mixture component | Final concn | Concn (μl)/reaction |
|---|---|---|
| Moltaq PCR-grade water | 13.575 | |
| Moltaq PCR buffer | 1× | 2.5 |
| dNTPs | 180 μM | 1.8 |
| Forward primer (785F) | 0.4 μM | 1 |
| Reverse primer (1175R) | 0.4 μM | 1 |
| Moltaq DNA polymerase | 25 mM | 0.125 |
| Template DNA | 5 | |
| Total | 25 |
dNTPs, deoxynucleoside triphosphates.
TABLE 7.
Cycling parameters for preparation of the library for 16S amplicon sequencing of placental samples
| Step | Temp (°C) | Time | No. of cycles |
|---|---|---|---|
| Initial denaturation | 94 | 3 min | 1 |
| Denaturation | 94 | 30 s | 32 |
| Primer annealing | 60 | 40 s | ↓a |
| Extension | 72 | 90 s | |
| Extension | 72 | 10 min | 1 |
The downward arrow indicates that denaturation, primer annealing, and 90-s extension were carried out over 32 cycles.
Identification of contaminants.
Negative extractions and PCR blanks from our study were examined for the presence of potential contaminants. Only negative samples with ≥500 reads were analyzed (n = 19). Any OTUs with at least two reads in at least two of the negative extraction samples were considered potential contaminating OTUs.
Bioinformatic analyses.
Paired-end 250-bp sequenced reads were merged by using FLASH v1.2.11. Reads were demultiplexed, quality filtered, and assigned taxonomic labels within the Quantitative Insights into Microbial Ecology (QIIME) 1.9.1 pipeline (50). The UCLUST algorithm (51) within QIIME was used to pick OTUs at 97% similarity against the Greengenes core reference database version 12.10 (52). Any sequences that failed to match at 97% similarity were clustered de novo by using UCLUST. De novo chimera removal was carried out by using UCHIME. A representative sequence was then chosen for each OTU, and this sequence was aligned to the Greengenes “Core Set” taxonomic alignment (53) by using PyNAST (54). These aligned sequences were used to build a phylogenetic tree by using FastTree 2.1.3 (55). Taxonomy was assigned by using RDP Classifier 2.2 (56) and the Greengenes taxonomy reference database, from which an OTU table was constructed.
Taxonomic labels were assigned to the highest possible level by using the QIIME pipeline, and data were converted to relative abundances for use in all subsequent analyses. The Phyloseq v1.22.3 package in R was used to combine OTU, clinical, taxonomic, and phylogenetic data into a single object suitable for relative-abundance comparisons and diversity analyses (57). Prior to analysis, samples with <500 reads were removed, as samples with low read depth do not capture the entire diversity of a sample, thus limiting the capacity to generate reliable diversity metrics.
To improve the power of regression models to identify differences in the placental microbiota between groups of interest, all biological replicates from both villous and parenchymal tissue were analyzed together in mixed-effect models that included a multilevel intercept for participant identifications. All analyses were carried out in R 3.4.3. Additional R packages and versions used are listed in the supplemental material.
Differential-abundance testing: Limma.
Raw abundance data were normalized by using the variance-stabilizing transformation (VST) approach in DESeq2 v1.18.1 (58) within Phyloseq. These normalized data were then transformed into a Limma v3.34.6 (59) object to utilize the multilevel functions available in this package. Any OTUs unassigned at the level of genus and with ≤10 reads were removed. Adjusted models were corrected for the potential confounding influences of delivery method, maternal ethnicity, collection hospital, maternal BMI, tissue type, and maternal smoking. All models were run with a random intercept to account for correlations between biological replicates. P values were corrected for multiple testing by using the Benjamini-Hochberg procedure to produce Q values (60). OTUs were also merged to the level of genus, and models were then rerun, to investigate genus-level, rather than OTU-level, associations.
Calculation of beta diversity.
Three common methods for assessing distance or dissimilarity between samples or groups of samples were performed by using the VST-normalized data of filtered OTU counts: weighted UniFrac (61), unweighted UniFrac (62), and the Bray-Curtis dissimilarity metric (63). VST matrices can include negative values, which represent zero or very low original counts and are not permitted by certain distance metrics. To mitigate this, any negative value was replaced with a zero, under the assumption that these cases were of very low, or near-zero, abundance and thus of negligible importance to the hypotheses under investigation. Following the computation of the three metrics, differences between the outcome groups of interest were visually explored by using PCoA. The adonis function in the Vegan v2.4.6 package (64) was used to quantify differences in beta diversity values between outcomes of interest. Significance tests were performed by using F-tests, from 999 permutations of the raw data.
Accession number(s).
Sequencing data have been deposited within the EMBL-EBI European Nucleotide Archive under study accession no. PRJEB25986.
Supplementary Material
ACKNOWLEDGMENTS
We thank Wellbeing of Women and Lord Mayor's funding. This research was funded by the United Kingdom Medical Research Council (MRC), the Biological Research Council, and Great Ormond Street Hospital Children's Charity (Ph.D. fellowship to L.J.L.) and was supported by the National Institute for Health Research Biomedical Research Centre at Great Ormond Street Hospital for Children, the NHS Foundation Trust, and University College London. P.S. is supported by Great Ormond Street Hospital Children's Charity and the MRC (MR/K022741/1 and MR/M004597/1). R.D. and N.K. were supported by the UCL Great Ormond Street Institute of Child Health and the Reuben Centre for Pediatric Virology and Metagenomics and by the Food and Nutrition Technical Assistance III Project (FANTA). T.G.C. is funded by the United Kingdom MRC (grant no. MR/K000551/1, MR/M01360X/1, MR/N010469/1, and MC_PC_15103). The MRC eMedLab computing resource was used for bioinformatics and statistical analyses.
We also thank all participating hospital staff and families.
We report no conflict of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00483-18.
REFERENCES
- 1.Khalil A, Rezende J, Akolekar R, Syngelaki A, Nicolaides KH. 2013. Maternal racial origin and adverse pregnancy outcome: a cohort study. Ultrasound Obstet Gynecol 41:278–285. doi: 10.1002/uog.12313. [DOI] [PubMed] [Google Scholar]
- 2.Muglia LJ, Katz M. 2010. The enigma of spontaneous preterm birth. N Engl J Med 362:529–535. doi: 10.1056/NEJMra0904308. [DOI] [PubMed] [Google Scholar]
- 3.Moser K, Stanfield KM, Leon DA. 2008. Birthweight and gestational age by ethnic group, England and Wales 2005: introducing new data on births. Health Stat Q 2008:22–31, 34–55. [PubMed] [Google Scholar]
- 4.Restrepo-Mendez MC, Lawlor DA, Horta BL, Matijasevich A, Santos IS, Menezes AM, Barros FC, Victora CG. 2015. The association of maternal age with birthweight and gestational age: a cross-cohort comparison. Paediatr Perinat Epidemiol 29:31–40. doi: 10.1111/ppe.12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shaw GM, Wise PH, Mayo J, Carmichael SL, Ley C, Lyell DJ, Shachar BZ, Melsop K, Phibbs CS, Stevenson DK, Parsonnet J, Gould JB. 2014. Maternal prepregnancy body mass index and risk of spontaneous preterm birth. Paediatr Perinat Epidemiol 28:302–311. doi: 10.1111/ppe.12125. [DOI] [PubMed] [Google Scholar]
- 6.Torloni MR, Betran AP, Daher S, Widmer M, Dolan SM, Menon R, Bergel E, Allen T, Merialdi M. 2009. Maternal BMI and preterm birth: a systematic review of the literature with meta-analysis. J Matern Fetal Neonatal Med 22:957–970. doi: 10.3109/14767050903042561. [DOI] [PubMed] [Google Scholar]
- 7.Girsen AI, Mayo JA, Carmichael SL, Phibbs CS, Shachar BZ, Stevenson DK, Lyell DJ, Shaw GM, Gould JB. 13 May 2016. Women's prepregnancy underweight as a risk factor for preterm birth: a retrospective study. BJOG doi: 10.1111/1471-0528.14027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldenberg RL, Culhane JF, Iams JD, Romero R. 2008. Epidemiology and causes of preterm birth. Lancet 371:75–84. doi: 10.1016/S0140-6736(08)60074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Onderdonk AB, Delaney ML, DuBois AM, Allred EN, Leviton A, Extremely Low Gestational Age Newborns Study Investigators . 2008. Detection of bacteria in placental tissues obtained from extremely low gestational age neonates. Am J Obstet Gynecol 198:110.e1–110.e7. doi: 10.1016/j.ajog.2007.05.044. [DOI] [PubMed] [Google Scholar]
- 10.Sykes L, MacIntyre DA, Yap XJ, Teoh TG, Bennett PR. 2012. The Th1:th2 dichotomy of pregnancy and preterm labour. Mediators Inflamm 2012:967629. doi: 10.1155/2012/967629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han YW, Shen T, Chung P, Buhimschi IA, Buhimschi CS. 2009. Uncultivated bacteria as etiologic agents of intra-amniotic inflammation leading to preterm birth. J Clin Microbiol 47:38–47. doi: 10.1128/JCM.01206-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hatanaka AR, Mattar R, Kawanami TE, França MS, Rolo LC, Nomura RM, Araujo E Jr, Nardozza LM, Moron AF. 16 December 2014. Amniotic fluid “sludge” is an independent risk factor for preterm delivery. J Matern Fetal Neonatal Med doi: 10.3109/14767058.2014.989202. [DOI] [PubMed] [Google Scholar]
- 13.Wang X, Buhimschi CS, Temoin S, Bhandari V, Han YW, Buhimschi IA. 2013. Comparative microbial analysis of paired amniotic fluid and cord blood from pregnancies complicated by preterm birth and early-onset neonatal sepsis. PLoS One 8:e56131. doi: 10.1371/journal.pone.0056131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. 2014. The placenta harbors a unique microbiome. Sci Transl Med 6:237ra65. doi: 10.1126/scitranslmed.3008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Onderdonk AB, Hecht JL, McElrath TF, Delaney ML, Allred EN, Leviton A. 2008. Colonization of second-trimester placenta parenchyma. Am J Obstet Gynecol 199:52.e1–52.e10. doi: 10.1016/j.ajog.2007.11.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doyle RM, Harris K, Kamiza S, Harjunmaa U, Ashorn U, Nkhoma M, Dewey KG, Maleta K, Ashorn P, Klein N. 2017. Bacterial communities found in placental tissues are associated with severe chorioamnionitis and adverse birth outcomes. PLoS One 12:e0180167. doi: 10.1371/journal.pone.0180167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fortner KB, Grotegut CA, Ransom CE, Bentley RC, Feng L, Lan L, Heine RP, Seed PC, Murtha AP. 2014. Bacteria localization and chorion thinning among preterm premature rupture of membranes. PLoS One 9:e83338. doi: 10.1371/journal.pone.0083338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones HE, Harris KA, Azizia M, Bank L, Carpenter B, Hartley JC, Klein N, Peebles D. 2009. Differing prevalence and diversity of bacterial species in fetal membranes from very preterm and term labor. PLoS One 4:e8205. doi: 10.1371/journal.pone.0008205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stout MJ, Conlon B, Landeau M, Lee I, Bower C, Zhao Q, Roehl KA, Nelson DM, Macones GA, Mysorekar IU. 2013. Identification of intracellular bacteria in the basal plate of the human placenta in term and preterm gestations. Am J Obstet Gynecol 208:226.e1–226.e7. doi: 10.1016/j.ajog.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Musilova I, Pliskova L, Kutova R, Hornychova H, Jacobsson B, Kacerovsky M. 28 November 2014. Ureaplasma species and Mycoplasma hominis in cervical fluid of pregnancies complicated by preterm prelabor rupture of membranes. J Matern Fetal Neonatal Med doi: 10.3109/14767058.2014.984606. [DOI] [PubMed] [Google Scholar]
- 21.de Andrade Ramos B, Kanninen TT, Sisti G, Witkin SS. 20 September 2014. Microorganisms in the female genital tract during pregnancy: tolerance versus pathogenesis. Am J Reprod Immunol doi: 10.1111/aji.12326. [DOI] [PubMed] [Google Scholar]
- 22.Han YW, Fardini Y, Chen C, Iacampo KG, Peraino VA, Shamonki JM, Redline RW. 2010. Term stillbirth caused by oral Fusobacterium nucleatum. Obstet Gynecol 115:442–445. doi: 10.1097/AOG.0b013e3181cb9955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prince AL, Ma J, Kannan PS, Alvarez M, Gisslen T, Harris RA, Sweeney EL, Knox CL, Lambers DS, Jobe AH, Chougnet CA, Kallapur SG, Aagaard KM. 2016. The placental membrane microbiome is altered among subjects with spontaneous preterm birth with and without chorioamnionitis. Am J Obstet Gynecol 214:627.e1–627.e16. doi: 10.1016/j.ajog.2016.01.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Charbonneau MR, Blanton LV, DiGiulio DB, Relman DA, Lebrilla CB, Mills DA, Gordon JI. 2016. A microbial perspective of human developmental biology. Nature 535:48–55. doi: 10.1038/nature18845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mysorekar IU, Cao B. 2014. Microbiome in parturition and preterm birth. Semin Reprod Med 32:50–55. doi: 10.1055/s-0033-1361830. [DOI] [PubMed] [Google Scholar]
- 26.Wassenaar TM, Panigrahi P. 27 October 2014. Is a fetus developing in a sterile environment? Lett Appl Microbiol doi: 10.1111/lam.12334. [DOI] [PubMed] [Google Scholar]
- 27.Payne MS, Bayatibojakhi S. 2014. Exploring preterm birth as a polymicrobial disease: an overview of the uterine microbiome. Front Immunol 5:595. doi: 10.3389/fimmu.2014.00595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kliman HJ. 2014. Comment on “The placenta harbors a unique microbiome.” Sci Transl Med 6:254le4. doi: 10.1126/scitranslmed.3009864. [DOI] [PubMed] [Google Scholar]
- 29.Perez-Muñoz ME, Arrieta M-C, Ramer-Tait AE, Walter J. 2017. A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: implications for research on the pioneer infant microbiome. Microbiome 5:48. doi: 10.1186/s40168-017-0268-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glassing A, Dowd SE, Galandiuk S, Davis B, Chiodini RJ. 2016. Inherent bacterial DNA contamination of extraction and sequencing reagents may affect interpretation of microbiota in low bacterial biomass samples. Gut Pathog 8:24. doi: 10.1186/s13099-016-0103-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF. 2014. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lauder AP, Roche AM, Sherrill-Mix S, Bailey A, Laughlin AL, Bittinger K, Leite R, Elovitz MA, Parry S, Bushman FD. 2016. Comparison of placenta samples with contamination controls does not provide evidence for a distinct placenta microbiota. Microbiome 4:29. doi: 10.1186/s40168-016-0172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rinke C, Low S, Woodcroft BJ, Raina J-B, Skarshewski A, Le XH, Butler MK, Stocker R, Seymour J, Tyson GW, Hugenholtz P. 2016. Validation of picogram- and femtogram-input DNA libraries for microscale metagenomics. PeerJ 4:e2486. doi: 10.7717/peerj.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kircher M, Sawyer S, Meyer M. 2011. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res 40:e3. doi: 10.1093/nar/gkr771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doyle RM, Alber DG, Jones HE, Harris K, Fitzgerald F, Peebles D, Klein N. 2014. Term and preterm labour are associated with distinct microbial community structures in placental membranes which are independent of mode of delivery. Placenta 35:1099–1101. doi: 10.1016/j.placenta.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 36.Combs CA, Gravett M, Garite TJ, Hickok DE, Lapidus J, Porreco R, Rael J, Grove T, Morgan TK, Clewell W, Miller H, Luthy D, Pereira L, Nageotte M, Robilio PA, Fortunato S, Simhan H, Baxter JK, Amon E, Franco A, Trofatter K, Heyborne K. 2014. Amniotic fluid infection, inflammation, and colonization in preterm labor with intact membranes. Am J Obstet Gynecol 210:125.e1–125.e15. doi: 10.1016/j.ajog.2013.11.032. [DOI] [PubMed] [Google Scholar]
- 37.DiGiulio DB. 2012. Diversity of microbes in amniotic fluid. Semin Fetal Neonatal Med 17:2–11. doi: 10.1016/j.siny.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 38.Hitti J, Riley DE, Krohn MA, Hillier SL, Agnew KJ, Krieger JN, Eschenbach DA. 1997. Broad-spectrum bacterial rDNA polymerase chain reaction assay for detecting amniotic fluid infection among women in premature labor. Clin Infect Dis 24:1228–1232. doi: 10.1086/513669. [DOI] [PubMed] [Google Scholar]
- 39.Pararas MV, Skevaki CL, Kafetzis DA. 2006. Preterm birth due to maternal infection: causative pathogens and modes of prevention. Eur J Clin Microbiol Infect Dis 25:562–569. doi: 10.1007/s10096-006-0190-3. [DOI] [PubMed] [Google Scholar]
- 40.Larsen B, Hwang J. 2010. Mycoplasma, Ureaplasma, and adverse pregnancy outcomes: a fresh look. Infect Dis Obstet Gynecol 2010:521921. doi: 10.1155/2010/521921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Capoccia R, Greub G, Baud D. 2013. Ureaplasma urealyticum, Mycoplasma hominis and adverse pregnancy outcomes. Curr Opin Infect Dis 26:231–240. doi: 10.1097/QCO.0b013e328360db58. [DOI] [PubMed] [Google Scholar]
- 42.Douvier S, Neuwirth C, Filipuzzi L, Kisterman J-P. 1999. Chorioamnionitis with intact membranes caused by Capnocytophaga sputigena. Eur J Obstet Gynecol Reprod Biol 83:109–112. doi: 10.1016/S0301-2115(98)00240-1. [DOI] [PubMed] [Google Scholar]
- 43.Lopez E, Raymond J, Patkai J, Ayoubl ME, Schmitz T, Moriette G, Jarreau PH. 2010. Capnocytophaga species and preterm birth: case series and review of the literature. Clin Microbiol Infect 16:1539–1543. doi: 10.1111/j.1469-0691.2010.03151.x. [DOI] [PubMed] [Google Scholar]
- 44.Hill GB. 1998. Preterm birth: associations with genital and possibly oral microflora. Ann Periodontol 3:222–232. doi: 10.1902/annals.1998.3.1.222. [DOI] [PubMed] [Google Scholar]
- 45.Campbell JR, Edwards MS. 1991. Capnocytophaga species infections in children. Pediatr Infect Dis J 10:944–948. [DOI] [PubMed] [Google Scholar]
- 46.Leon LJ, Solanky N, Stalman SE, Demetriou C, Abu-Amero S, Stanier P, Regan L, Moore GE. 2016. A new biological and clinical resource for research into pregnancy complications: the Baby Bio Bank. Placenta 46:31–37. doi: 10.1016/j.placenta.2016.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuan SQ, Cohen DB, Ravel J, Abdo Z, Forney LJ. 2012. Evaluation of methods for the extraction and purification of DNA from the human microbiome. PLoS One 7:e33865. doi: 10.1371/journal.pone.0033865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Boer R, Peters R, Gierveld S, Schuurman T, Kooistra-Smid M, Savelkoul P. 2010. Improved detection of microbial DNA after bead-beating before DNA isolation. J Microbiol Methods 80:209–211. doi: 10.1016/j.mimet.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 49.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 52.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A. 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. 2010. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McMurdie PJ, Holmes S. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Methodol 57:289–300. [Google Scholar]
- 61.Lozupone CA, Hamady M, Kelley ST, Knight R. 2007. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol 73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bray JR, Curtis JT. 1957. An ordination of the upland forest communities of southern Wisconsin. Ecol Monogr 27:326–349. doi: 10.2307/1942268. [DOI] [Google Scholar]
- 64.Dixon P. 2003. VEGAN, a package of R functions for community ecology. J Veg Sci 14:927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.