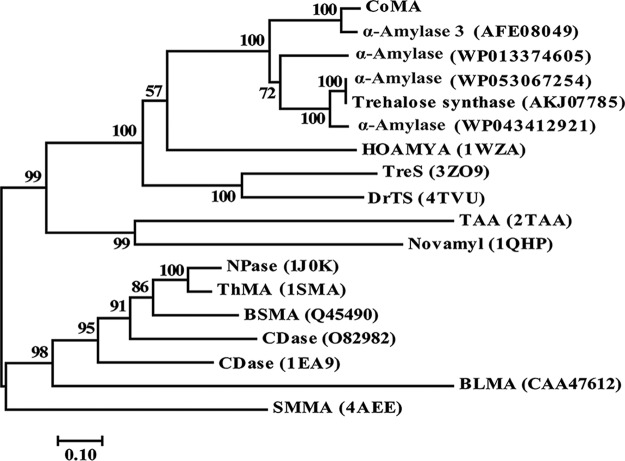
FIG 1.
Phylogenetic relationships between CoMA and other amylases from GH13. The phylogenetic tree was constructed by the neighbor-joining algorithm based on the amino acid sequence alignment in MEGA7. The amino acid sequence of CoMA was aligned with those of the following proteins: C. coralloides DSM 2259 α-amylase 3 (accession no. AFE08049), Stigmatella aurantiaca α-amylase (WP013374605), Archangium gephyra α-amylase (WP053067254), Archangium gephyra trehalose synthase (AKJ07785), Cystobacter violaceus α-amylase (WP043412921), Halothermothrix orenii α-amylase (HOAMYA; PDB 1WZA), Mycobacterium smegmatis trehalose synthase (TreS; PDB 3ZO9), Deinococcus radiodurans trehalose synthase (DrTS; PDB 4TVU), Aspergillus oryzae Taka-amylase A (PDB 2TAA), Bacillus stearothermophilus Novamy (PDB 1QHP), Bacillus stearothermophilus neopullulanase (NPase; PDB 1J0K), Thermus sp. IM6501 maltogenic amylase (ThMA; PDB 1SMA), Bacillus sp. A2-5a cyclomaltodextrinase (CDase; O82982), Bacillus sp. CDase (PDB 1EA9), Bacillus licheniformis maltogenic amylase (BLMA; CAA47612), and Staphylothermus marinus maltogenic amylase (SMMA; PDB 4AEE).