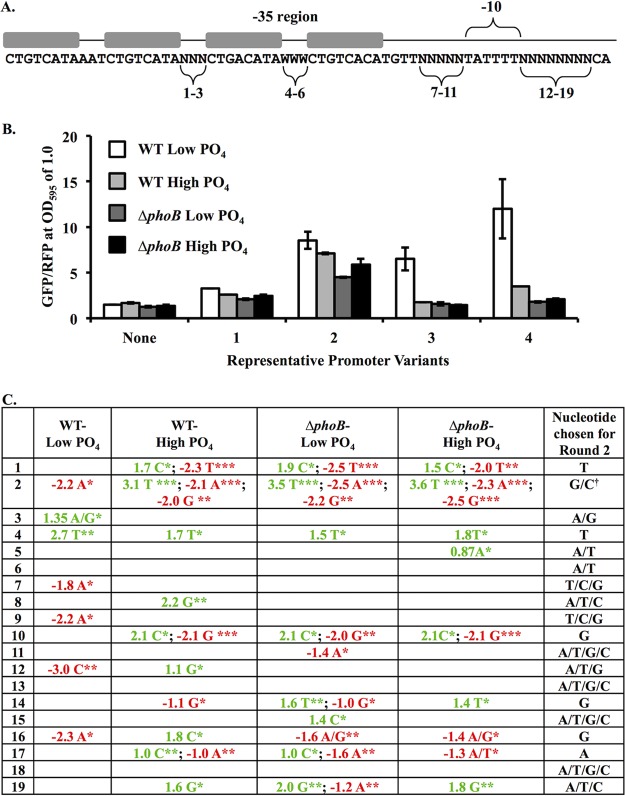
FIG 1.
Design and screening of synthetic PhoB-dependent promoter variants. (A) Synthetic constructs included four PhoB-binding sites (gray boxes) as well as a −10 promoter element. The −35 promoter region is indicated, although it does not resemble a −35 consensus. Randomized or semirandomized positions are numbered, corresponding to the numbering in panel C. “N” indicates a randomized position (labeled 1 to 3 and 7 to 19). Positions 4 to 6 were restricted to A or T (W). Eighty-three variants of the sequence were cloned upstream of gfp and screened for activity. (B) GFP output for four representative promoter variants. GFP was measured for constructs in ES114 and JLS9 (ΔphoB) grown in FMM medium with a high (378 μM) or low (37.8 μM) added PO4 concentration. Strains with the promoterless parent vector (labeled “None”) show background fluorescence. Values were taken at an OD595 of 1.0 and normalized to red fluorescence. Error bars indicate standard deviations (n = 3). Data from one representative experiment of three are shown. (C) Each variable position (positions 1 to 19) was subjected to a Mann-Whitney U nonparametric test to compare the average GFP output associated with each individual nucleotide (A, T, C, or G) against the average value for constructs with the other nucleotides in that position. Pairs of nucleotides were also compared at each position (A/T versus G/C, C/T versus A/G, G/T versus A/C, and C/G versus A/T). Only comparisons with P values of <0.05 are shown. Values indicate the mean GFP output for all constructs with the nucleotide (or nucleotide pair) at the indicated position minus the mean value for the other constructs. For each of the four strain-medium combinations, nucleotides that correlated with significantly higher or lower GFP levels are indicated by green or red, respectively; *, **, and *** indicate P values of <0.05, <0.01, and < 0.001, respectively. The right column indicates how randomization was further constrained for the next set of variants (round 2). † indicates that the C at position 2 was underrepresented in the screened variants (in only 1 of the 83 variants) and therefore was included in round 2.