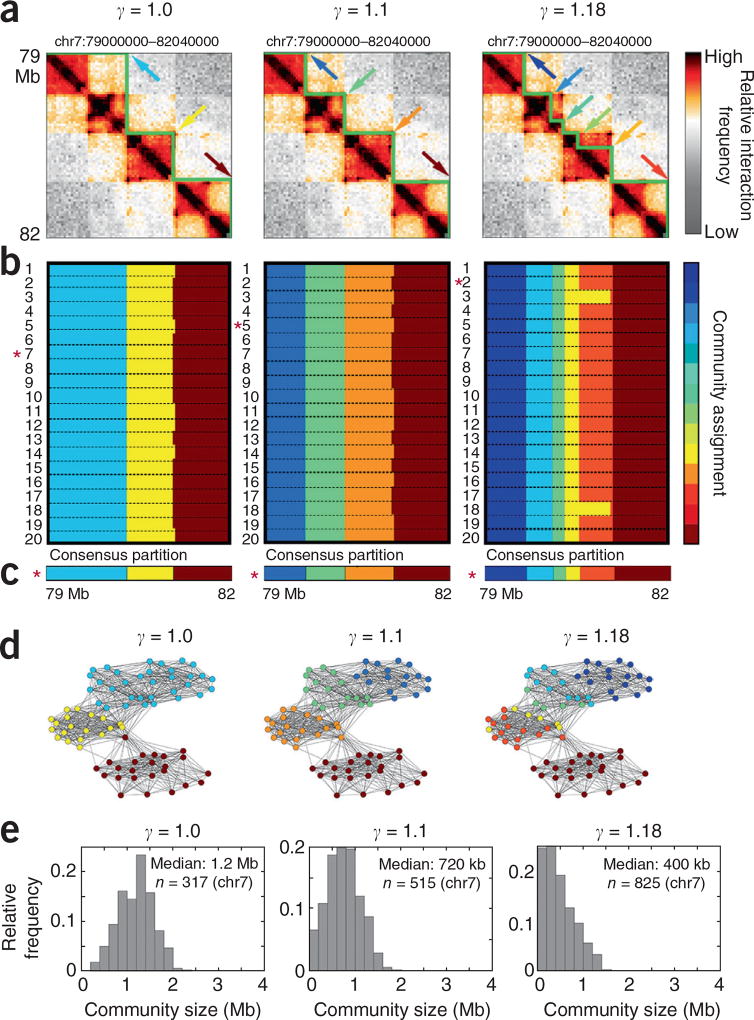
Figure 1.
Network modularity maximization and consensus partitioning (3DNetMod-MMCP) identifies nested, partially overlapping chromatin domains across length scales. (a) Hi-C heatmaps from human cortical plate tissue16. Domains identified with 3DNetMod-MMCP at γ = 1 (left), γ = 1.1 (center) and γ = 1.18 (right) are outlined in green. (b) Community partitions from 20 applications of 3DNetMod-MM at γ = 1 (left), γ = 1.1 (center) and γ = 1.18 (right). (c) Consensus partitions for each γ value from 20 partitions. (d) Spring-force diagrams representing network communities in Hi-C data. Node proximity corresponds to the relative interaction frequency between the two genomic bins. Node color indicates community assignment. (e) Chromosome-wide distribution (chr7) of domain sizes identified with γ = 1.0 (left), γ = 1.1 (center) and γ = 1.18 (right).