Figure 1. Detecting somatic mosaicism in the genome through various sequencing strategies.
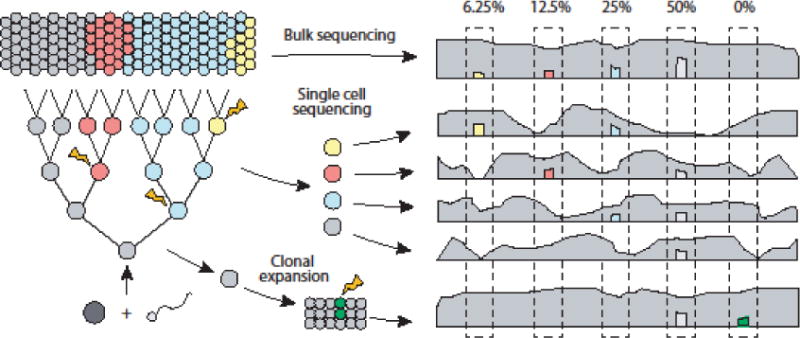
Somatic mutations arise during development and propagate to a sub-population of cells (blue: 50% of cells; red: 25%; yellow: 12.5%). With bulk sequencing, these somatic mutations are expected to be approximately half of the sub-population frequency. Lower frequency somatic mutations require higher sequencing depth to maintain detection sensitivity. With single cell sequencing, somatic mutations can be detected as heterozygous variants that occur in a subset of cells. The ability to detect variants is dependent on uniformity of coverage and allelic balance in genome amplification, as well as picking cells that contain variants. Clonal expansion followed by bulk sequencing does not suffer from the problems associated with single cell sequencing, but artifactual mutations that occur early during expansion (green) can be difficult to distinguish from mutations in the original cell.
