Figure 9.
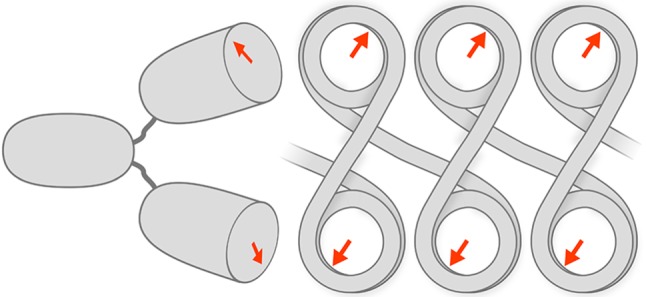
Cartoon sketch depicting a bivalent PL2-6 molecule binding to a zig-zag arrangement of nucleosomes. This sketch does not display accurate molecular dimensions. Rather, it is exaggerated to show the potential sites of interaction. A more accurate model can be visualized in the electronic supplementary material, Movie. The cartoon arrows on the nucleosome ‘faces’ represent the nucleosome acidic patches (AP). Notice that in this zig-zag arrangement, for sequential nucleosomes, the AP orientations are related by a dyad axis (perpendicular to the linker DNA and to the plane of the cartoon). Adjacent to the zig-zag chromatin is a representation of the bivalent PL2-6, also displaying the complementary binding regions as arrows. A bivalent IgG has a dyad axis running between the two Fab regions and along the midline of the Fc region. The junction between the Fab and Fc regions is very flexible, tolerating a wide range of angles between the Fab ‘arms’ and allowing the arms to rotate with considerable freedom. The geometry of the nucleosomes presents two binding sites to the bivalent IgG, exploiting the avidity principle.
