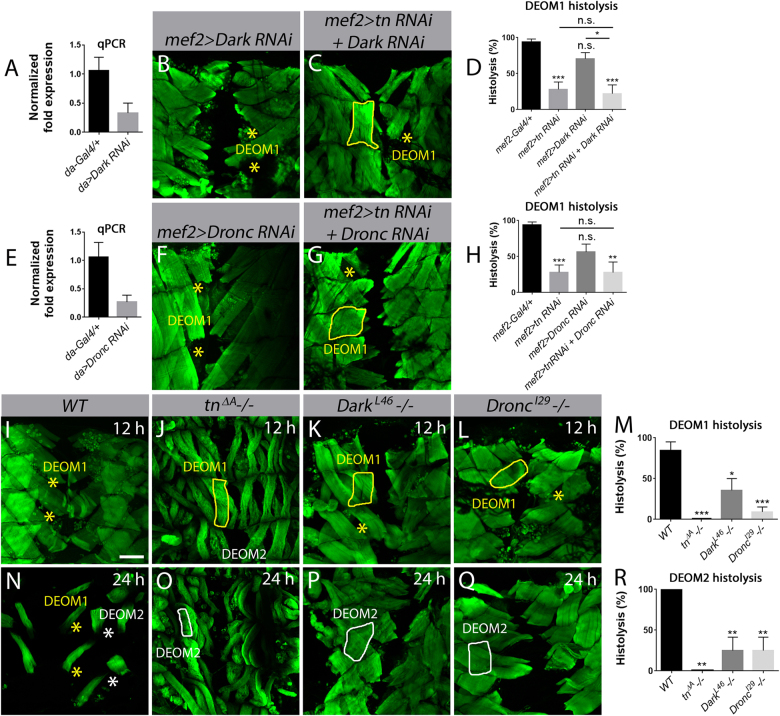
Fig. 5. Dronc and Dark are required for DEOM histolysis.
Merged Z-stack confocal images of the abdominal muscles stained with phalloidin (green) at 12 h APF (b, c, f, g, i–l) or 24 h APF (n–q). DEOM1s are outlined with a solid yellow line, DEOM2s with a solid white line. Histolyzed DEOM1 and DEOM2 muscles are marked by yellow and white asterisks, respectively. a Dark transcript levels are reduced by over 50% using the ubiquitous da-Gal4 driver. N = 3 biological replicates and 3 technical replicates for each genotype. b, c DEOM1 histolysis is partially blocked upon RNAi knockdown of Dark alone (b) or if Dark is reduced in a tn RNAi background at 12 h APF. d Quantification of DEOM1 histolysis reveals a slight enhancement in muscle degeneration in mef2>tn RNAi+Dark RNAi compared to mef2>Dark RNAi alone. e Quantification of Dronc transcript levels show over a 60% decrease upon expression of UAS-Dark RNAi under control of da-Gal4. N = 3 biological replicates and 3 technical replicates for each genotype. f–h There is no significant difference in the histolysis of DEOM muscles in mef2>tn RNAi+Dronc RNAi pupae compared to Dronc RNAi. i–l WT DEOM1s have histolyzed by 12 h APF (i). There is a strong block in muscle breakdown in tnΔA (j), DarkL46 (k), and DroncI29 (l) homozygous mutants at this same time point. m A bar graph showing the quantification of DEOM1 histolysis in (i–l). n–q All DEOM muscles are absent in WT pupae at 24 h APF (n). tnΔA (o), DarkL46−/− (p), and DroncI29−/− (q) show significantly reduced DEOM2 breakdown at 24 h APF. r Quantification of DEOM2 histolysis in tnΔA, DarkL46, and DroncI29 mutants. Mean ± SEM (n.s., not significant, ***p < 0.005, **p < 0.01, *p < 0.05). Scale bar, 100 µm (b, c, f, g, i–l, n–q)