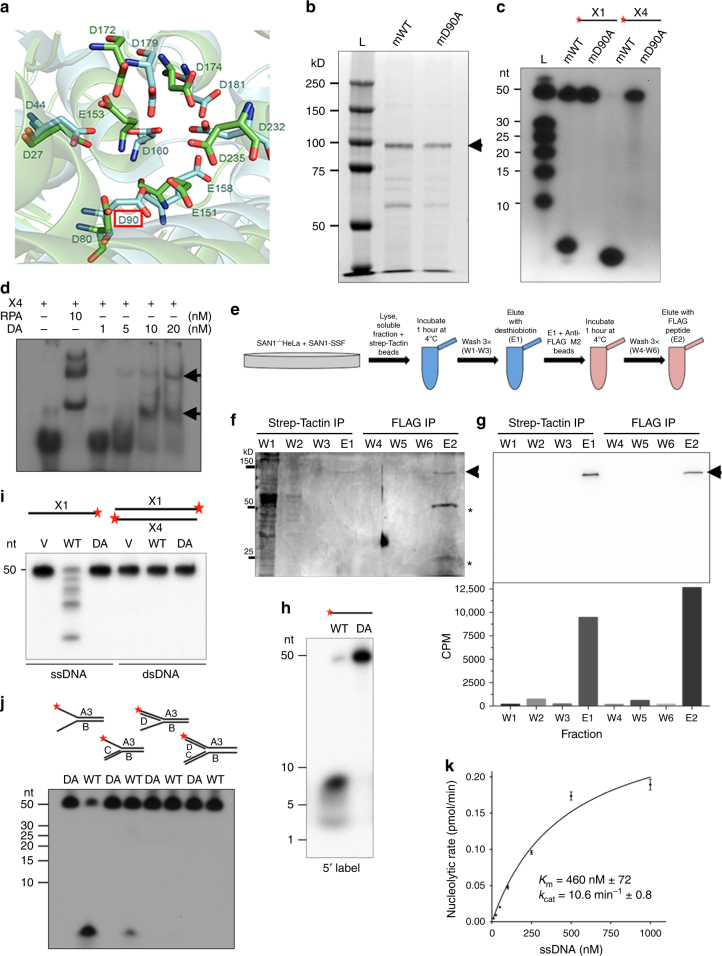
Fig. 1.
Identification and characterization of the SAN1 nuclease. a Modeling of conserved carboxylates in active site of SAN1 (light blue), using the Robetta server (http://robetta.bakerlab.org) and the A. fulgidus FEN1 structure (PDB 1RXW) (green) as template. Residue highlighted by red box is the aspartate mutated to make D90A. b Murine SAN1 was expressed with a C-terminal Strep tag in E. coli and purified over Strep-Tactin beads. Purified protein (0.2 µg) was analyzed by PAGE and stained with Coomassie. Arrow shows mSAN1 expected size of 100 kD. c Synthetic 50-mer oligos were 5′ labeled with 32P and incubated with mSAN1 for 120 min. Products were separated by PAGE and 32P-fragments were detected by autoradiography (see Table 1 for sequences). d 50 nt X4 was 5′ 32P labeled and incubated with RPA as a positive control or increasing concentrations of SAN1 D90A (catalytically inactive). Samples were analyzed on a native gel and exposed to X-ray film. e Schematic of double-affinity Strep-FLAG tag purification for human SAN1 WT and SAN1 DA. f Silver stained fractions from the purification where “W” denotes Wash steps and “E” denotes Elution steps for the Strep and FLAG IPs. Arrow shows human SAN1 WT (expected size 150 kD) and asterisks show FLAG antibody heavy and light chains. g Top panel shows immunoblot of fractions from two-step purification of SAN1 where arrow shows SAN1 (expected size 150 kD), detected using mouse M2 anti-FLAG-antibody. Bottom panel shows corresponding filter spin nuclease assay. h X1 (50-mer ssDNA) was 5′ labeled with 32P and incubated with SAN1 WT or the D90A mutant. Products were analyzed as in c. i X4 ssDNA or dsDNA X1 + X4 were 3′ 32P labeled and incubated with WT or D90A SAN1. Products were analyzed as in h. j FLAG-tagged SAN1 WT or D90A was incubated with 5′ 32P labeled splayed duplex, 3′ flap, or 5′ flap structures for 2 h at 37 °C. Products were processed as in c. k Using the filter spin assay, initial rates of 5′ 32P-labeled X4 hydrolysis were measured at different substrate concentrations. Line was fitted using Prism software, assuming Michaelis-Menten kinetics