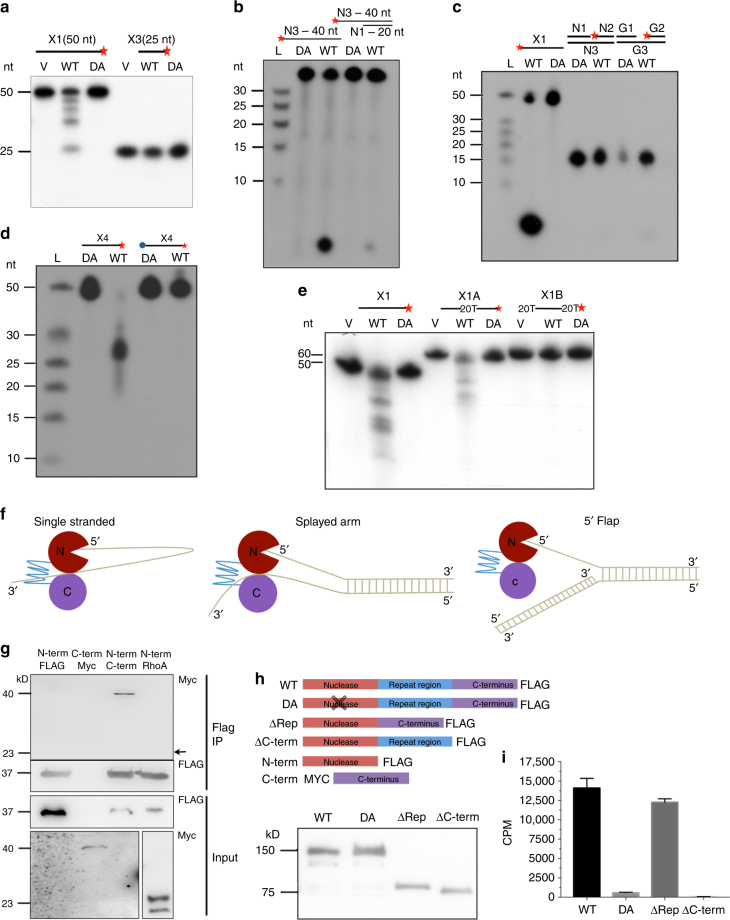
Fig. 2.
SAN1 acts as a 5′ exonuclease on single-stranded DNA substrates. Affinity-purified FLAG-SAN1 WT or -D90A from 293T cells incubated with a 50 nt ssDNA versus 25 nt ssDNA; b 40 nt ssDNA versus a 20 nt ssDNA 5′ overhang followed by 20 bp of dsDNA; c dsDNA oligos with an internal nick or gap; d 5′ biotinylated X4 and unbiotinylated X4; e variants of ssDNA oligo X1 with 20 Ts 5′ and 3′ to 20 nts of X1, or a tract of 20 Ts bounded by two 20 nt sections of the X1 sequence all for 2 h at 37 °C. DNA structures assayed in b, c were 5’ 32P labeled and structures in a, d, e were 3′ 32P labeled. Products were separated by PAGE and 32P fragments were detected by autoradiography. f Schematic of a model for SAN1 nuclease activity. SAN1 acts on ssDNA substrates by recognizing the free 5′ end of DNA and cleaving ~3 or ~7 nts from the 5′ end, in a non-processive manner. g The N-terminal FLAG-tagged nuclease domain was co-expressed in 293T cells with C-terminal Myc-tagged SAN1 C-terminus, or Myc-RhoA as a negative control. Lysates were precipitated with anti-FLAG M2 beads and analyzed by immunoblot. The Myc Input membrane was re-exposed for a longer time to detect Myc-SAN1 C-terminus. The SAN1 C-terminal domain but not RhoA is co-precipitated with the nuclease domain. h Schematic of SAN1 deletion mutants used in f, g, h followed by immunoblot of WT, DA, ΔRep, and ΔC-term proteins purified from 293T cells; and i tested for nuclease activity against 5′ 32P labeled ssDNA using the filter spin assay (N = 2, error bars show range)