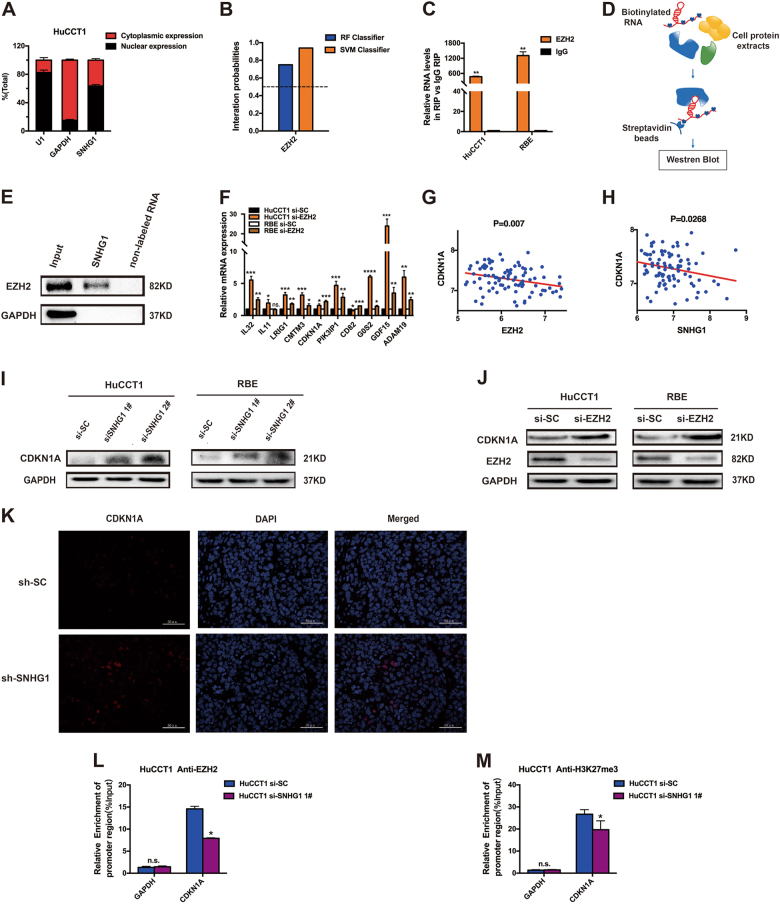
Fig. 6. SNHG1 binds with EZH2 in the nucleus, thus leading to epigenetical silencing of CDKN1A.
a After nuclear and cytosolic separation, RNA expression levels were measured by qRT-PCR. GAPDH was used as a cytosol marker and U1 was used as a nucleus marker. b The interation probabilies of EZH2 and SNHG1 were detected on the website, and the results showed that EZH2 could well bind with SNHG1 well (http://pridb.gdcb.iastate.edu/RPISeq/index.html). c RIP experiment for EZH2 was performed and the co-precipitated RNA was subjected to qRT-PCR for SNHG1. d Schematic of the RNA pull-down experiment for the identification of proteins associated with SNHG1. e In vitro transcribed, pull-down assays showed that desthiobiotinylated SNHG1 could retrieve EZH2 in HuCCT1 cells, but not GAPDH. GAPDH was a negative control. f The methylation-related genes were detected by qRT-PCR in HuCCT1 and RBE cell lines after knockdown of EZH2. g The correlation between EZH2 and CDKN1A expression was detected by analyzing GSE76297 data. h The correlation between SNHG1 and CDKN1A expression was detected by analyzing GSE76297 data. i The altered protein levels of CDKN1A were selectively confirmed by western blot in knockdown of SNHG1. j The altered protein level of CDKN1A was selectively confirmed by western blot in knockdown EZH2. k Immunofluorescence was done to explore if the expression of CDKN1A was changed by knockdown of SNHG1 in vivo. l, m ChIP of EZH2 and H3K27me3 of the promoter region of CDKN1A locus after siRNA treatment targeting si-SC and si-SNHG1 1# in HuCCT1 cells; qPCR was performed to detect the quantitation of ChIP assays. Enrichment was quantified relative to input controls. Antibody directed against IgG was used as a negative control. Error bars indicate means ± SD. *P < 0.05; **P < 0.01;***P < 0.001;****P < 0.0001; n.s. not significant.