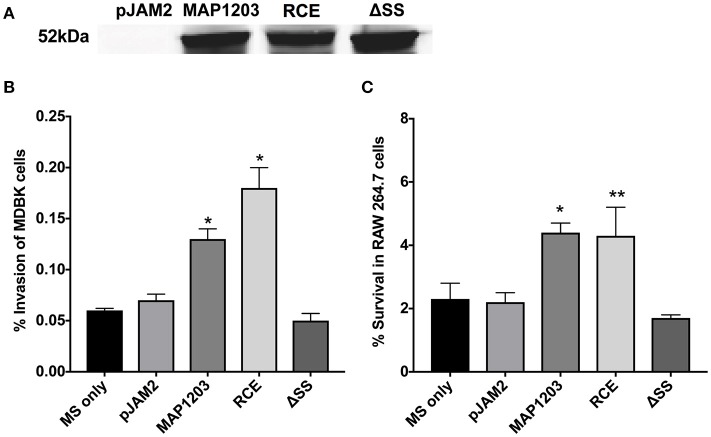
Figure 2.
MAP1203 overexpression in M. smegmatis. (A) Cultures of M. smegmatis clones containing the control empty plasmid pJAM2, pJAM2:MAP1203, pJAM2:RCE, or pJAM2:ΔSS were grown to an OD600 0.5 and then induced by adding 0.2% acetamide and 0.2% succinic acid to the medium for 6 h at 37°C at 200 rpm. Bacteria were mechanically disrupted in the bead-beater and cleared lysates were separated on the SDS-PAGE gel for Western blotting of the MAP1203 protein (52 kDa). The blot shown is representative of the expression of constructs for all M. smegmatis assays completed. (B) MDBK epithelial cells were infected with the induced clones of M. smegmatis for 2 h and percentage of invasion was calculated from the original inoculum used to infect the bovine cells. *p < 0.05, significance compared to M. smegmatis with and without pJAM2 vector. (C) The RAW 264.7 macrophages were incubated with the induced clones of M. smegmatis and after 2 h extracellular bacteria were removed from cells by washing and antibiotic treatment. After 14 h of infection, macrophages were lysed and intracellular bacterial were quantified by Colony Forming Unit counts. The percentage of survival was calculated from the CFU counts of intracellular bacteria that entered RAW 264.7 cells during 2 h invasion. Data represent the mean ± SEM of three independent experiments conducted in triplicate. *p < 0.05, **p < 0.01 significance compared to M. smegmatis with and without pJAM2 vector as determined by Student's t-test.