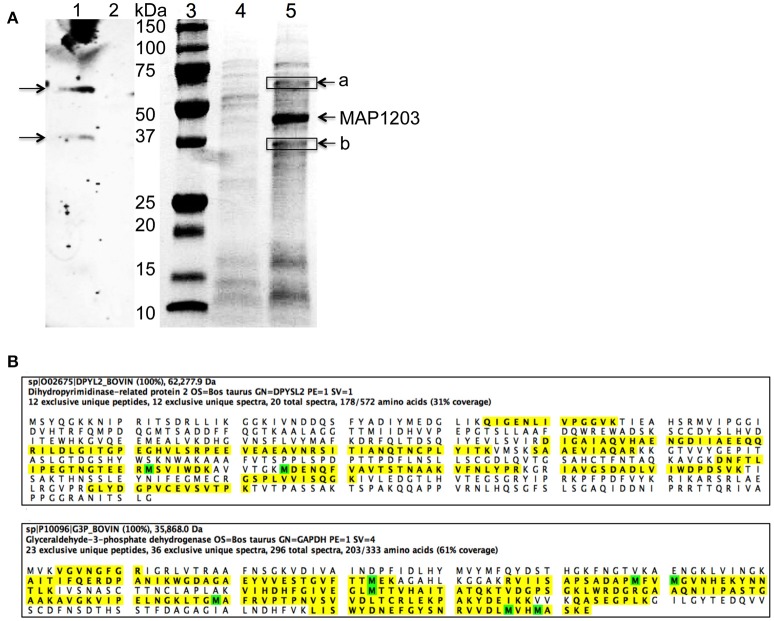
Figure 5.
The host proteins interacting with MAP1203. (A) Identification of potential MAP1203 binding host partners by far-Western blotting and pull-down assays. (Lane 1) The biotin-labeled surface proteins of MDBK cells were transferred to a nitrocellulose membrane and incubated with the recombinant MAP1203 (0.5 mg) overnight at 4°C on the shaker. Bound proteins were visualized with 6xHN antibody as described in the materials and methods; (Lane 2) The control membrane (without any host proteins) was subjected to same procedures as lane 1; (Lane 3) Molecular Weight, Protein molecular marker. (Lane 4) The control pull-down assay was performed with E. coli lysate that contained the empty overexpression vector, and was exposed to total proteins of MDBK cells. Bands visualized with Coomassie staining represent non-specific host proteins that were bound to the column; (Lane 5) The cleared sample of E. coli overexpressing MAP1203 protein was incubated with the total protein lysate of bovine epithelial cells and unique proteins identified in the experimental lane (a and b) were excised and sequenced. (B) The MAP1203 bound host proteins were identified as Dihydropyrimidinase-related protein 2 and Glyceraldehyde 3-phosphate dehydrogenase; the identified peptides by MS are marked in yellow; Oxidized Methionine (M) is marked in green.