Figure 5. STK11/LKB1 mutations are a genomic determinant of poor clinical outcome with PD-1 axis blockade in PD-L1 positive non-squamous NSCLC, regardless of KRAS status.
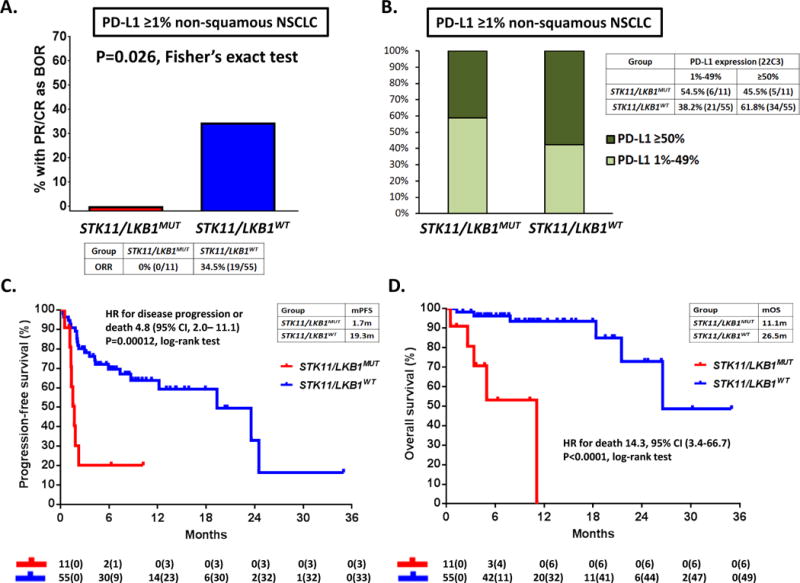
A. Objective response rate (RECISTv1.1) to PD-1/PD-L1 inhibitors in STK11/LKB1-mutant and wild-type patients with PD-L1-positive non-squamous NSCLC (≥1%) from MDACC (N=66). PD-L1 expression was assessed using the FDA-approved 22C3 pharmDx assay (Dako). A two-tailed Fisher’s exact test (computed from a 2×2 contingency table) was used to assess the significance of the association between group membership (STK11/LKB1-mutant versus STK11/LKB1 -wild-type) and best overall response (PR/CR vs SD/PD). B. Fractions of PD-L1 low-positive (1%-49%) and PD-L1 high-positive (≥50%) tumors in the STK11/LKB1-mutant and wild-type groups. C. Kaplan-Meier estimates of progression-free survival with PD-1/PD-L1 blockade in STK11/LKB1-mutant and wild-type groups. Tick marks represent data censored at the last time the patient was known to be alive and without disease progression (date of last radiological assessment). D. Kaplan-Meier estimates of overall survival with PD-1 inhibitors in the STK11/LKB1-mutant and wild-type groups. Tick marks represent data censored at the last time the patient was known to be alive.
