Figure 2.
Defects in Arc Ubiquitination Enhance mGluR-LTD
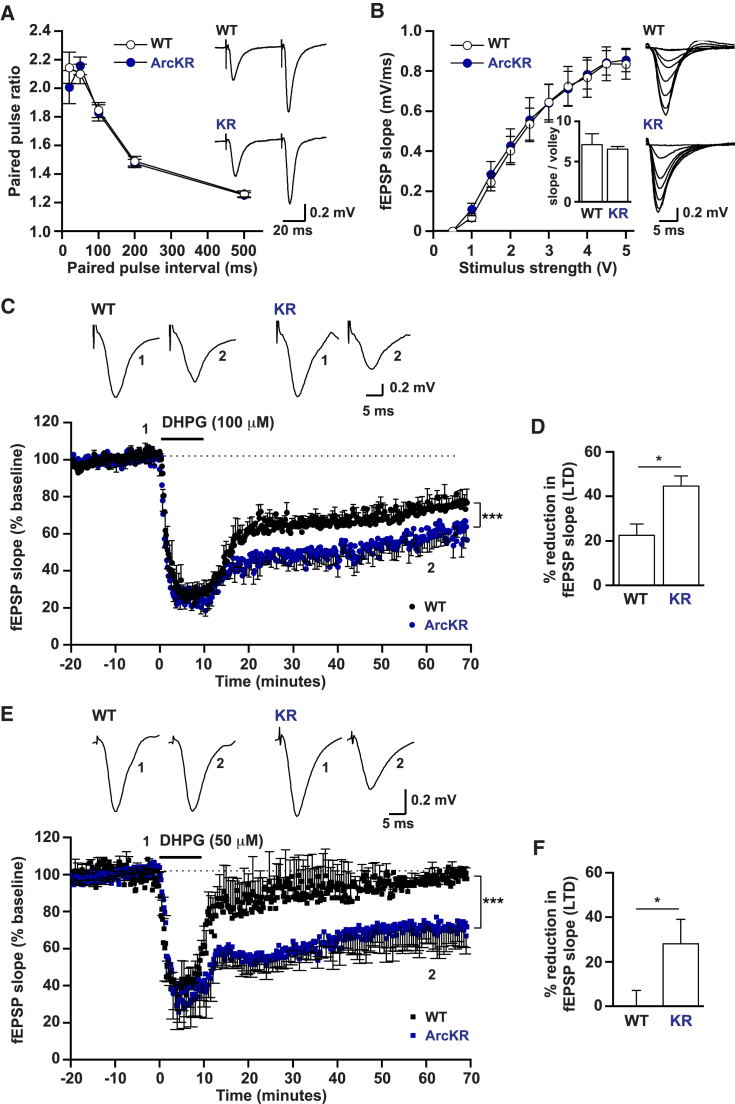
(A) Mean paired-pulse ratio against paired-pulse interval for WT (n = 12 slices) and ArcKR mice (n = 10 slices). Inset: representative traces at an interval of 50 ms from WT and ArcKR littermates.
(B) Mean fEPSP slope against stimulus strength for WT (n = 9 slices) and ArcKR mice (n = 10 slices) with examples of superimposed averaged fEPSPs at different stimulus strengths. Inset: ratio of fEPSP slope versus volley amplitude at 40% of the stimulus strength that gives the maximum fEPSP slope (n = 6 slices per genotype).
(C) Normalized mean fEPSP slope against time for WT and ArcKR mice. After a 20 min baseline, DHPG (100 μM) was applied for 10 min and then washed out for 1 hr. Baseline fEPSP slope was analyzed at 15–20 min and LTD was analyzed at 55–60 min after DHPG application (fEPSP slope was significantly reduced following DHPG, ArcKR, p = 0.00016; WT, p = 0.00013, fEPSPs were not normalized). LTD was significantly enhanced in ArcKR slices versus WT littermates (WT: 74.3% ± 3%, n = 5 animals, 9 slices; ArcKR: 49.5% ± 4.3%, 5 mice, 12 slices, p = 0.0004, fEPSPs were normalized to baseline). fEPSP traces (averages of 10 fEPSPs) were taken at the times indicated by the numerals in the plot below.
(D) Mean percentage reduction in fEPSP slope (LTD) between 55 to 60 min after DHPG application (p = 0.015).
(E) Normalized mean fEPSP slope against time for WT and ArcKR mice. Slices were treated as in (C). In ArcKR slices, fEPSP slope was significantly reduced after 55–60 min of 50 μM DHPG application (p = 0.0046) but was not significantly (p = 0.41) reduced in WT slices (fEPSPs were not normalized). fEPSP traces (averages of 10 fEPSPs) were taken at the numerals in the lower plot.
(F) LTD was not induced in WT mice by 50 μM DHPG (reduction in slope 1.8% ± 7.7%, n = 3 mice, 5 slices) but was induced in ArcKR littermates (reduction 28% ± 11%, 3 mice, 5 slices, p = 0.04). Values represent mean ± SEM. Statistical comparisons were carried out with one-way ANOVA, paired and unpaired Student’s t tests.