Abstract
Simultaneously analysing morphological, molecular and stratigraphic data suggests a potential resolution to a major remaining inconsistency in crocodylian evolution. The ancient, long-snouted thoracosaurs have always been placed near the Indian gharial Gavialis, but their antiquity (ca 72 Ma) is highly incongruous with genomic evidence for the young age of the Gavialis lineage (ca 40 Ma). We reconcile this contradiction with an updated morphological dataset and novel analysis, and demonstrate that thoracosaurs are an ancient iteration of long-snouted stem crocodylians unrelated to modern gharials. The extensive similarities between thoracosaurs and Gavialis are shown to be an almost ‘perfect storm’ of homoplasy, combining convergent adaptions to fish-eating, as well resemblances between genuinely primitive traits (thoracosaurs) and atavisms (Gavialis). Phylogenetic methods that ignore stratigraphy (parsimony and undated Bayesian methods) are unable to tease apart these similarities and invariably unite thoracosaurs and Gavialis. However, tip-dated Bayesian approaches additionally consider the large temporal gap separating ancient (thoracosaurs) and modern (Gavialis) iterations of similar long-snouted crocodyliforms. These analyses robustly favour a phylogeny which places thoracosaurs basal to crocodylians, far removed from modern gharials, which accordingly are a very young radiation. This phylogenetic uncoupling of ancient and modern gharial-like crocs is more consistent with molecular clock divergence estimates, and also the bulk of the crocodylian fossil record (e.g. all unequivocal gharial fossils are very young). Provided that the priors and models attribute appropriate relative weights to the morphological and stratigraphic signals—an issue that requires investigation—tip-dating approaches are potentially better able to detect homoplasy and improve inferences about phylogenetic relationships, character evolution and divergence dates.
Keywords: homoplasy, convergence, Bayesian phylogenetics, tip-dating, crocodiles, archosaurs
1. Introduction
Among living crocodiles and alligators (Crocodylia), the most extreme feeding specializations are shown by gharials (gavials): the Indian gharial Gavialis gangeticus and the Malayan or ‘false’ gharial Tomistoma schlegelii. Both have similar trophic structures: highly elongate, narrow snouts with retracted nares, and slender, sharp, regularly spaced, uniform-sized teeth. The evolution and biogeography of these fascinating and endangered reptiles have been heavily studied (e.g. [1–4]). Systematists long interpreted their similarity as convergence for fish-eating (e.g. [5–7]), and morphological data alone continues to separate the two living gharials (figure 1). Tomistoma groups with ‘true’ crocodiles (Crocodylinae: Crocodylus and Osteolaemus); Gavialis, in contrast, is basal to all other living crocodylians (Alligatoridae + Crocodylidae). The fossil record was also interpreted as supporting this arrangement. Many narrow-snouted fossils extending as far back as the Mesozoic were found to be related to Gavialis, i.e. stem-gharials [5]. The earliest of these proposed stem-gharials are thoracosaurs, a group of early crocodylians with elongate, narrow snouts that are found mostly in Late Cretaceous to Early Paleogene marginal marine deposits of Europe and North America [7]. The oldest thoracosaurs are ca 72 Ma, almost as old as the oldest undisputed crown crocodylians [9], thus suggesting the early divergence of the Gavialis lineage.
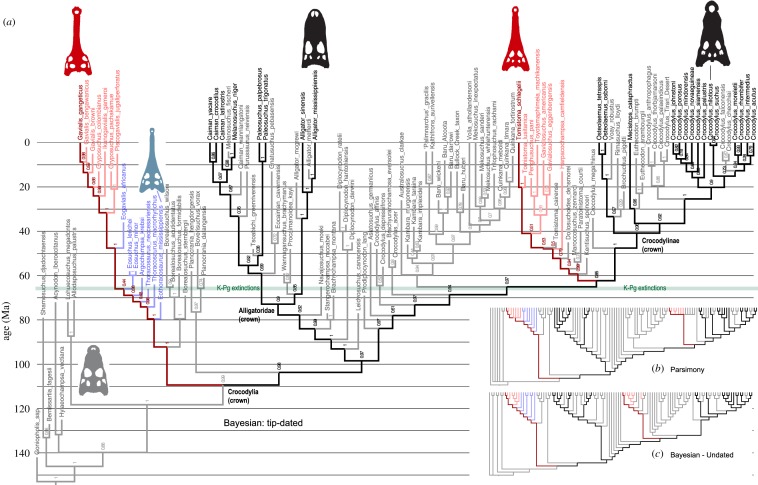
Figure 1.
Morphology-only phylogeny of crocodilians based on (a) tip-dated Bayesian, (b) parsimony and (c) undated Bayesian approaches. All methods yield similar trees, in which Gavialis is sister to all other living crocodilians, which is strongly contradicted by molecular and combined analyses. The long-snounted, ancient thoracosaurs are always robustly placed on the lineage leading to Gavialis. Dark fonts and branches denote living taxa; lighter fonts and branches denote extinct taxa. Living gharials (Gavialis and Tomistoma) are dark red, fossil crown gharials (figure 2) are light pink; thoracosaurs are light blue. Numbers in (a) refer to clade posterior probabilities; see [8, figures S1–S3] for detailed versions.
By contrast, analyses of large molecular datasets—alone or in combination with morphology—have consistently supported an arrangement where Indian and Malayan gharials are sister taxa, forming an expanded Gavialidae (hereafter, ‘gharials’: figure 2). In this topology, gharials and crocodylids (now shorn of Tomistoma) form a larger clade (Longirostres), while alligatorids are now the sister group of all other crocodylians. The Gavialis–Tomistoma clade is very strongly supported by numerous mitochondrial and nuclear genes [1,2,10,11], and also consistent with certain morphological traits [1,12]. This arrangement implies that the extreme fish-eating specializations of Indian and Malayan gharials are homologous rather than convergent. Furthermore, molecular dating (using calibrations outside gharials) strongly suggests that the Gavialis–Tomistoma divergence is very recent (ca 40 Ma: [10]), with crown crocodylians as a whole being only ca 70 Ma [13].
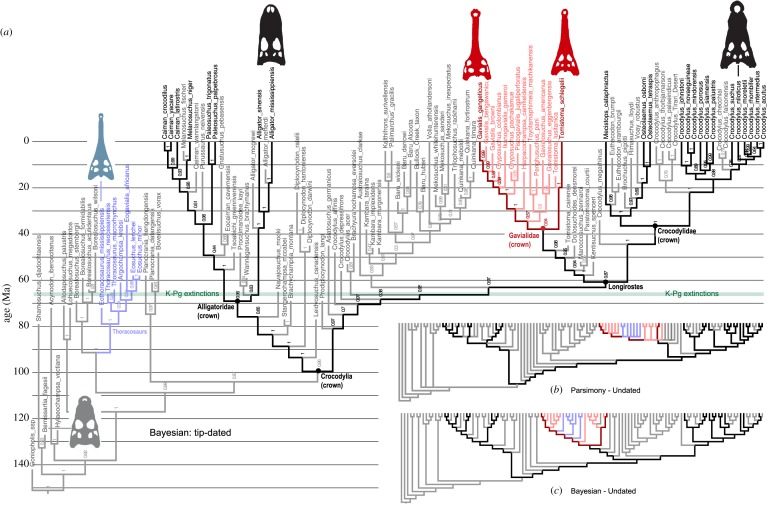
Figure 2.
DNA + morphology phylogeny of crocodilians based on (a) tip-dated Bayesian, (b) parsimony and (c) undated Bayesian approaches. All methods unite living gharials (Gavialis + Tomistoma); however, tip-dating robustly places the long-snounted, ancient thoracosaurs as basal forms outside all living taxa, while parsimony and undated Bayesian analyses place them robustly within the gharial clade. Dark fonts and branches denote living taxa; lighter fonts and branches denote extinct taxa. Living gharials (Gavialis and Tomistoma) are dark red, fossil crown gharials (as identified in Fig. 2a) are light pink; thoracosaurs are light blue. Numbers in (a) refer to clade posterior probabilities; see [8, figures S4–S6] for detailed versions.
However, while the emerging consensus of a gharial (Gavialis–Tomistoma) clade helps reconcile the molecular and anatomical data, there are still some major contradictions. In combined analyses of DNA and morphology where this gharial clade is strongly retrieved, the fossil thoracosaurs continue to emerge on the stem lineage of Gavialis, and thus within crown gharials (e.g. [1,12]). However, the age of thoracosaurs (up to ca 72 Ma) is highly inconsistent with the young age of crown gharials as independently estimated by molecular data (less than 40 Ma) using a small number fossil calibrations, and suggested by the bulk of the gharial fossil record (all other gharial fossils are less than 20 Ma). Indeed, the oldest thoracosaurs substantially pre-date the oldest unequivocal fossils of the more-inclusive clade Longistrostes. The position of thoracosaurs within gharials thus not only contradicts molecular divergence estimates—which might be biased by the few selected calibration points—but also, apparently, much of the crocodylian fossil record.
This stratigraphic incongruence strongly suggests that thoracosaurs might be basal or stem crocodylians which convergently evolved the elaborate feeding specializations of Gavialis. Furthermore, Gavialis—uniquely among living crocodylians—has evolved a range of atavistic (secondarily primitive) traits [1]. Thus, many of the primitive-looking traits in thoracosaurs, which might typically lead them to be placed as basal crocodylians, could also be interpreted as atavisms further linking them to Gavialis. The problematic grouping of thoracosaurs and Gavialis thus potentially represents one of the most intractable cases of homoplasy, involving extensive convergent adaptation, as well as similarities shared due to plesiomorphy and atavism.
Here, we show that new phylogenetic methods that take into account stratigraphy (i.e. the discordant ages of thoracosaurs and gharials) in the context of both morphological and molecular data are uniquely able to untangle this intricate web of homoplasy. Building on earlier studies, we present an expanded matrix of the anatomy of fossil and living crocodylians including both crown and stem forms (i.e. Crocodyliformes sensu [14]), and combine this with multi-locus genetic data for all living species. Analyses of this comprehensive dataset using parsimony and undated Bayesian methods continue to group thoracosaurs with modern gharials. However, tip-dated total-evidence phylogenetics, which treat fossils as time-series samples of phenotypes, are uniquely able to tease apart the two separate waves of evolution of gharial-like forms. The large temporal gap separating these instances of convergent evolution leads thoracosaurs to be placed outside of all living crocodylians, thus separating them from the much younger modern gharials. This potentially resolves a major remaining conflict in crocodylian phylogeny: the ancient occurrence of ‘gharial-like’ forms (revealed by morphology and stratigraphy) but apparent short history of living gharials (strongly supported by genomics). Additional corroboration could come from new fossils with unusual character combinations consistent with this interpretation. Tip-dating phylogenetic methods are able to detect homoplasy by harnessing statigraphic information in combination with molecular and morphological data. They are thus not only a promising approach for time-scaling phylogenies, but may also improve estimates of topological relationships and character evolution.
2. Material and Methods
(a). Morphological, molecular and stratigraphic data
(i). Morphological and stratigraphic data
One hundred and seventeen crocodylian taxa (25 living and 92 extinct) were scored for 278 morphological characters, expanding the character and taxon sampling from [6] (66 taxa and 166 characters). A full list of characters and their sources is provided in the Dryad Digital Repository [8, appendix 1], as are the sources (literature and specimens) for the scores given to newly added characters and taxa. Stratigraphic information and sources are also in the Dryad Digital Repository [8, DataAndResultFiles.zip].
(ii). Molecular data
A molecular supermatrix for all 25 living taxa in our phylogeny, with 9284 base pairs consisting of exons and introns from 10 nuclear gene loci, and three mtDNA loci with associated tRNAs and DLoop, was constructed from published data ([1,10] and GenBank). Full details are in the Dryad Digital Repository [8, appendix 1].
(b). Analyses
A full description of all analyses can be found in the Dryad Digital Repository [8, appendix 2], along with executable files containing the morphological, molecular and combined matrices in nexus, MrBayes and BEAST formats (DataAndResultsFiles.zip). The analyses are summarized below.
The morphological data were analysed alone, and in combination with the molecular data (mor + mol), using parsimony (PAUP* [15]), undated Bayesian (MrBayes [16]) and tip-dated Bayesian (BEAST including BEASTMC3 [17]) methods, yielding a total of six primary analyses. The first two analytic approaches estimated topology and anagenetic branch lengths only. The tip-dated analyses co-estimated topologies, branch lengths (anagenetic and chronological), divergence dates and evolutionary rates [18,19]. Root age or more specifically the start of the diversification process was given an essentially uninformative prior of 0–1000 Ma and no internal node-age priors were imposed; the retrieved dates are thus largely driven by the phenotypic and stratigraphic information contained in the fossil taxa (tips). In the undated and dated Bayesian analyses, the optimal partitioning scheme and substitution models for the molecular data were chosen by PartitionFinder [20], and for the morphological data (i.e. with or without gamma) using stepping-stone methods. In the dated analyses, the optimal clock model (strict or relaxed) for the morphological data, and for molecular data, were also chosen via stepping-stone methods. All Bayesian analyses were repeated four times to test for stationarity, with the post-burn-in samples of all four runs combined for statistical analyses and consensus trees. Tracer [21] and AWTY [22] were used to ascertain burn-in and parameter convergence for numerical and topology, respectively.
The phylogenetic position of the seven thoracosaurs remained unstable across different analyses of the combined mor + mol data: they emerged within gharials when stratigraphic information was not considered (parsimony and undated Bayesian: figure 2b,c), but were a basal lineage outside of all living crocodylians when their antiquity was considered (tip-dated Bayesian: figure 2a). This suggests that the stratigraphic age of thoracosaurs is an important factor that helps separate them from gharials. To test this, a further tip-dated Bayesian analysis of the mor + mol data was performed after removing the stratigraphic information in the seven thoracosaurs (i.e. their stratigraphic ages were changed to a flat prior between 0 and ∞). Furthermore, the position of thoracosaurs within gharials appeared to suggest anomalously deep divergence dates across the entire crocodylian tree, compared to the remaining (i.e. non-thoracosaur) crocodylian fossil record (see Results and discussion). To test whether the two suites of fossils (thoracosaur and non-thoracosaur) imply fundamentally incompatible divergence dates, additional tip-dated analyses of the mor + mol performed with (i) living taxa and all non-thoracosaur fossils and (ii) living taxa and only thoracosaurs.
Morphological support for the two radically different positions for thoracosaurs was also evaluated using PAUP, to identify the conflicting suites of characters favouring affinities with gharials or a basal position.
Finally, to test whether our results might be susceptible to the effects of character ordering, character state definitions and taxon sampling (all of which inevitably are partly subjective in morphological analyses), we repeated the combined morphological and molecular analysis using an alternative morphological dataset which employed a different character and taxon sampling, and treated multistate characters as all unordered [23]. As the key phylogenetic conclusions were the same (see [8, figure S9]), our discussion focuses on the analyses using the dataset presented here.
3. Results and discussion
(a). The short history of long-snouted crocodylians
Analyses of the morphological data alone using all methods (parsimony, undated and tip-dated Bayesian) retrieved similar trees for living and fossil taxa: the Indian gharial Gavialis was basal to all other living crocodylians, while the Malayan gharial Tomistoma was closely related to crocodylines (figure 1). Thoracosaurs were always on the stem leading to Gavialis (bootstrap BS = 80%, Bayesian PP = 1.0), consistent with the historical hypothesis of the early divergence and long history of Gavialis. These results are highly consistent with previous parsimony analyses of morphology (e.g. [5–7]). However, the arrangement of living forms in this tree has now been strongly refuted by molecular data (see Introduction), so the remainder of the discussion will focus on analyses of the combined morphological and molecular data.
Analyses of the combined (mor + mol) data yielded similar results across all methods regarding the interrelationships of living taxa (figure 2). Gavialis and Tomistoma are closest living relatives (together: gharials), and their fish-eating specializations are thus homologous rather than convergent. Gharials are related to crocodylids, and alligatorids are sister to all other living crocodylians. These arrangements were stable across all methods and also consistent with all recent analyses that incorporate substantial genetic data (e.g. [1,2,10,11,13]). That the interrelationships of living taxa would conform to the molecular data was expected due to the relatively larger molecular dataset (greater than 9000 versus less than 300 characters) and the strength and cohesion of the molecular signal.
The positions of most fossil taxa were also stable in mor + mol analyses, across all methods. However, the affinities of the gharial-like thoracosaurs are variable, with the alternative positions have major stratigraphic and evolutionary implications. In parsimony and undated Bayesian analyses (figure 2b,c)—which ignored their stratigraphic antiquity—they emerged within crown gharials, on the stem lineage leading to Gavialis (BS = 69%, PP = 0.88). This is consistent with previous (parsimony) mor + mol analyses [1,12]. Given that the oldest thoracosaurs are Cretaceous in age, this position pushes crown gharials (and all nodes below this) deep into the Mesozoic. This greatly exceeds molecular clock estimates for the age of crown gharials (ca 40 Ma). Furthermore, the stratigraphic ages of most fossil crocodylians (excluding thoracosaurs) are also generally consistent with the shallow molecular divergences: crown crocodylians are almost exclusively Cenozoic, with only a handful of basal taxa known from the Late Cretaceous, and unequivocal fossil gharials are also younger than 20 Ma (see below). In parsimony and Bayesian analyses which infer undated topologies and thus do not consider stratigraphic information, these chronological anomalies are not considered, and the morphological similarities between thoracosaurs and Gavialis are taken as evidence of a relationship.
The tip-dated Bayesian analyses robustly retrieved a very different (and more stratigraphically consistent) position for thoracosaurs, placing them outside of gharials and in fact remote from all living crocodylians (PP = 0.97: figure 2a). Tip-dating strongly supports relatively shallow divergences across all living crocodylians, retrieving a young (ca 40 Ma) age for crown gharials: almost identical to conventional molecular clock studies with node-age constraints based on other crocodylian fossils. Along with thoracosaurs, other anomalously old, putative ‘tomistomines’ (e.g. Kentisuchus spenceri and Maroccosuchus zennaroi, ca 50 Ma) are also excluded from the Gavialis–Tomistoma clade. There are thus far fewer fossil taxa remaining in crown gharials, and all are very young. The sole fossil taxon on the lineage leading to living Tomistoma is Tomistoma lusitanica (ca 18 Ma). Several fossils fall on the lineage leading to living Gavialis, with the oldest being Penghusuchus (ca 16 Ma). The enigmatic Harpacochampsa camfieldensis (ca 13 Ma) from Australia emerges as another relative of living Gavialis, and not a mekosuchine (e.g. [24]). Notably, when rates of morphological evolution are assessed across crocodylians, the fastest branch is within gharials (figure 3): these fast rates suggest relatively simultaneous acquisition of adaptations for piscivory along with atavistic changes (reversals) on the lineage leading to Gavialis, which occurred between 22 and 28 Ma.
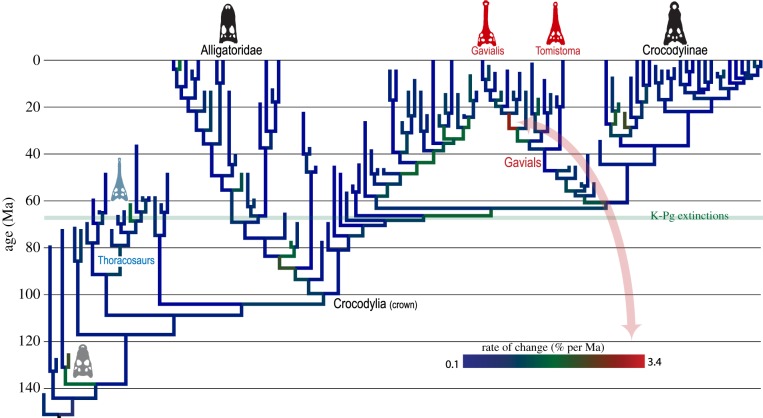
Figure 3.
Rates of morphological evolution in crocodilians; warmer colours denote faster rates. Rates are quite uniform through time and across clades, but the fastest occurs on a recent branch (22–27 Ma) leading to Gavialis, when adaptations to piscivory, and atavistic reversals, occurred relatively simultaneously. This branch thus generates a gharial morphotype highly similar to thoracosaurs, but these iterations are widely separated in time. See [8, figure S7] for precise branch values.
Tip-dating, unlike time-free methods, thus strongly retrieves living gharials as a young clade with an accordingly short fossil record. The body plan of Gavialis, in particular, appeared recently and relatively suddenly, via the most rapid rates of evolution with crocodylians.
(b). Tip-dating and the temporal signal of convergence
Tip-dated Bayesian analyses co-estimate topology and divergence dates. The extensive molecular and morphological data generate a robust tree and relative divergences between living crocodylians, and this tree is anchored in time by the morphology and ages of the fossil taxa (tips). The bulk of the fossil tips (non-thoracosaurs) places shallow absolute dates on this tree, whereas the thoracosaurs tend to cluster with Gavialis and thus tries to place deeper dates on this tree. However, only one of these absolute timescales can be correct. When all taxa are considered, the larger suite of crocodylian fossils carries the day (figure 2a): the molecular tree is calibrated with shallow divergence dates, and the ancient thoracosaurs cannot cluster within gharials (as they pre-date this clade). They emerge in a basal position—the position in this dated chronogram most consistent with their morphology and stratigraphic age.
These points were highlighted by three additional tip-dated analyses of the mor + mol data. The first evaluated where thoracosaurs would best ‘fit’ based on morphology alone (i.e. in the absence of stratigraphic data), and also what stratigraphic ages would be most consistent with this preferred position. The tip-dated analysis was repeated but the stratigraphic age of thoracosaurs (only) was changed to ‘unknown’ (flat prior 0 to ∞) and allowed to be estimated. Without stratigraphic constraints, thoracosaurs emerged within gharials (on the Gavialis lineage) with strong support (PP = 1), mirroring the results of the undated analyses [8, figure S8]. However, the stratigraphic ages implied by this position were very young: 5–15 Ma compared with actual ages of 36–72 Ma. This emphasizes the incongruity between the Gavialis-like morphology of thoracosaurs, and their early stratigraphic appearance, which leads to tip-dating approaches favouring two convergent events that are widely separated in time.
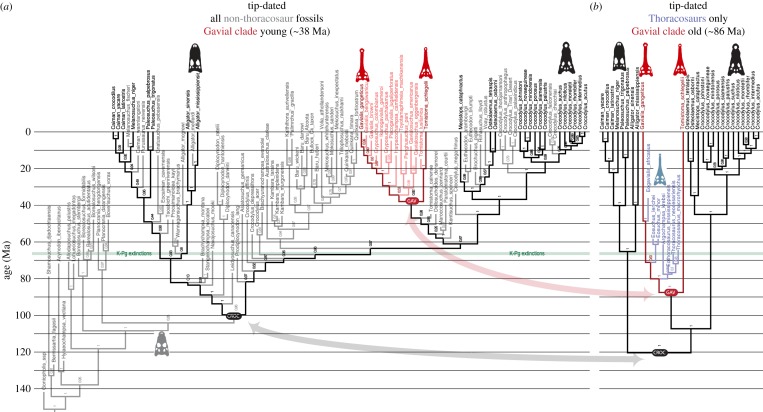
The second two additional analyses identified the discordant temporal signals suggested by the bulk of the crocodylian fossil record (excluding thoracosaurs), and by thoracosaurs, and also demonstrated how the majority signal prevails. The tip-dated analysis of the full mor + mol data was compared to analyses of two reduced datasets: (a) living taxa and all non-thoracosaur fossils and (b) living taxa and only thoracosaurs. Analysis (a) produced a tree with near-identical topology and divergence dates to the full dataset (e.g. gharials are ca 38 Ma in both, crocodylians are ca 100 Ma: figures 3 and 4a). Analysis (b), however, again retrieved thoracosaurs inside gharials, with inflated divergence dates for gharials (ca 87 Ma) and crocodylians as a whole (ca 120 Ma): figure 4b. Interestingly, in a combined analysis of all taxa, the differing temporal signals of the non-thoracosaur and thoracosaur fossils are not ‘averaged’ to give intermediate divergence dates (because such dates would fit neither suite of fossils). Rather, the majority non-thoracosaur signal strongly prevails and shallow dates are retrieved; the stratigraphically aberrant fossils do not distort (deepen) divergence dates, but shift to a more basal position where they no longer cause a temporal problem.
Figure 4.
Highly contradictory temporal signals are exhibited by thoracosaurs and the rest of the crocodilian fossil record: DNA + Morphology phylogenies of crocodilians, tip-dated using only non-thoracosaur fossils and only thoracosaurs. (a) Non-thoracosaur fossils only, resulting in shallow divergences similar to those retrieved by the full dataset (figure 2a). (b) Thoracosaurs only, resulting in very different and much deeper divergences. Colour coding as in figure 2; numbers refer to clade posterior probabilities.
(c). Tip-dating, tree topology and identification of homoplasy
The strong impacts of tip-dating on topology substantially influences associated inferences, such as divergence dates and character evolution. These effects might be considered as a glass that is either ‘half full’ or ‘half empty’; we first discuss them in a potentially positive light, but then temper this with important caveats.
Thoracosaurs and Gavialis, if indeed not closely related, represent one of the most intractable examples of homoplasy: a complex combination of convergent adaptations for aquatic feeding, as well as shared similarities that are genuinely primitive (in thoracosaurs) and atavistic (in Gavialis). It is not surprising that analyses of the anatomical data alone (parsimony and undated Bayesian methods)—even in concert with molecular data—have consistently united these taxa (e.g. [1,5–7,12]). However, a very ancient single origin of gharials is highly inconsistent with the bulk of the crocodylian stratigraphic record, as well as molecular divergence data. Tip-dating methods consider the wide temporal gap separating these similar taxa, and favours two separate iterations of gharial-like forms: the hypothesis most consistent with morphological, molecular and stratigraphic data considered together is that the plesiomorphic traits in thoracosaurs are genuinely primitive (shared with other basal crocodylians rather than gharials), while the jaw specializations are an ancient, convergent iteration of the gharial condition.
The two different potential positions of thoracosaurs represent diametrically different interpretations of their jaw adaptations and plesiomorphic traits. Affinities with gharials can potentially explain most of the jaw adaptations, and some of the plesiomorphies, as homologies with gharials (e.g. [7]); a basal position implies convergence in jaw anatomy with gharials, but more readily explains most plesiomorphic traits (as genuine primitive retentions). When the relative support for these two hypotheses is quantified by optimizing morphology on these two trees, 56 characters favour nesting with gharials, but 36 characters favour a basal position (see [8, appendix 2]). In particular, thoracosaurs appear to retain an ossified epipterygoid (described in Eosuchus minor [25], not scoreable in others), and tall posterior pterygoid processes, while lacking a caudal laterosphenoid bridge connecting to the quadrate: these plesiomorphies are found in other stem crocodylians but are largely or totally absent in crown gharials (Gavialis, Tomistoma and their undisputed fossil relatives). Thus, the basal position of thoracosaurs, while not being the optimal on purely morphological grounds, nevertheless has some intriguing supporting plesiomorphies, and is preferred in the tip-dated analysis because it is much more consilient with the bulk of the stratigraphic record of crocodylians and molecular divergence dates.
In this instance, the fortuitously wide temporal gap separating successive iterations of the gharial-like crocodylians allows tip-dating to untangle extensive homoplasy. However, this will not always be the case: for instance, if the thoracosaur lineage survived until ca 20 Ma and only then evolved their Gavialis-like traits, there would be less stratigraphic inconsistency with erroneously placing these long-snouted forms with Gavialis. The survival to the present of one lineage (Gavialis) is also critical, allowing accurate molecular assessment of its phylogenetic affinities and thus the recency of its phenotypic traits. It is notable that tip-dated analyses of crocodylians without molecular data (figure 1 and [26]) still link thoracosaurs and Gavialis. A truly perfect storm of homoplasy would involve ancient and totally extinct taxa (beyond even the reach of ancient DNA), which evolve similar adaptations at similar times; such problems might be intractable even to tip-dated total-evidence analyses.
Tip-dating methods have usually been discussed in the context of time-scaling trees (e.g. [18,26–28]). However, they can also improve estimates of topology by better identifying homoplasy—especially when that homoplasy is temporally separated. Tip-dating co-estimates topology, divergence dates, evolutionary rates and other related parameters, and in this instance disfavours a topology which although favoured by the morphological data (even in the context of a molecular backbone: figure 2b,c) is highly inconsistent with the temporal data (figure 4b).
(d). Caveats and conclusions
The topological effects of tip-dating, discussed in a positive light above, might alternatively be viewed as disconcerting. As an extreme example, a fossil taxon which is older than all other taxa in a phylogenetic analysis, but totally unknown for all characters, could be placed anywhere in a parsimony analysis, but would tend to be recovered in basal positions in a tip-dated analysis. Either result could be argued to be more ‘reasonable’: the character data are consistent with any position (in accordance with parsimony), but if forced to predict the fossil's true position, most systematists would guess a basal position is more likely (in accordance tip-dating). Similarly, it is widely accepted that phylogenies can be independently tested against the geological record (e.g. [29]). However, if one is prepared to choose among equally-parsimonious trees using stratigraphic concordance (e.g. [30]), what happens if a topology only a single step longer is even more stratigraphically concordant [31]? Analogously, it could be argued that keeping stratigraphy separate is advisable because such data are fundamentally different from morphology, and also allows stratigraphy to be independent test of tree topology. Alternatively, it could be argued that incorporating stratigraphy in phylogenetic reconstruction could result in globally optimal solutions that could not be discovered by considering these data sources sequentially.
Approaches to incorporate stratigraphy into phylogenetic reconstruction have a long but chequered history, with ‘stratocladistics’ being a recent quantitative, parsimony-based development [32]. However, that approach appears to have gained only limited acceptance. While there are many issues of contention [32], of relevance here are problems integrating character and stratigraphic data. In stratocladistics, each extra inferred ghost lineage (regardless of duration) is typically treated as exactly equivalent to one extra step of morphological homoplasy, but it is unclear whether these two sources of evidence can be equated in such a simplistic manner. A Bayesian framework provides a more mathematically precise common currency: impact on posterior probability. Trees with more homoplasy but greater stratigraphic concordance will only be preferred if there is a net positive impact on posterior probability. Importantly, these relative impacts are evaluated in the context of a complex adopted model of evolutionary change, which incorporates more biologically relevant aspects than a stratocladistic model, e.g. DNA and morphological substitution and clock models, absolute (rather than relative) fossil ages as well as their uncertainties, lineage sampling frequency, speciation and extinction rates, node-age priors, etc. While model testing approaches are now widely used to refine some of these parameters (see [8, appendix 2]), other model aspects are typically less well tested (e.g. sampling and diversification), and their overall interactions are even less explored. But the specifics of the overall model can potentially strongly influence the relative ‘balance’ between the morphological and stratigraphic data even though they are expressed in a common currency, and it remains to be tested whether widely used implementations (such as the models used here) are affording insufficient, adequate or exaggerated weight to the stratigraphic data.
Finally, the topological effects of tip-dating highlight the impact of simultaneously, rather than sequentially, considering data from multiple sources. Simultaneous, tip-dating approaches retrieve a dated phylogeny of crocodylians highly consistent with molecular data and the bulk of the crocodylian fossil record, and with the pleisiomorphies of thoracosaurs: thoracosaurs as basal forms and shallow divergences between all living lineages. However, the common sequential approach of estimating an undated topology first (ignoring stratigraphy), and then time-scaling it afterwards, would produce highly anomalous results. A highly suspect undated topology—where thoracosaurs fall within gharials—would be retrieved in the first step. Time-scaling this topology would lead to a dated chronogram with deep divergence dates that contradicts not only most molecular analyses, but also most of the crocodylian fossil record: any phylogeny of crocodylians where the Gavialis–Tomistoma divergence is greater than 72 Ma old would imply improbably long ghost lineages for a large number of crocodylian lineages. A simultaneous, rather than sequential, approach to the thoracosaur–gharial problem offers a more globally consistent solution to this long-standing problem in crocodylian evolution. Other instances of suspected homoplasy across temporally separated taxa could be approached in a similar manner.
Acknowledgements
We thank Jamie Oaks for discussion, and John Gatesy, Chris Brochu and Peter Makovicky for extensive comments which greatly improved the paper, though all errors and misconceptions remain ours alone.
Data accessibility
Full details of the methods and results, along with datasets and analysis executables, have been deposited in the Dryad Digital Repository (http://dx.doi.org/10.5061/dryad.3d23687) [8].
Authors' contributions
A.M.Y. collected most of the data, with assistance of M.L. M.L. performed most of the analyses, with assistance of A.M.Y. Both authors wrote the paper.
Competing interests
We declare we have no competing interests.
Funding
This study was funded by Australian Research Council grants to M.S.Y.L.
References
- 1.Gatesy J, Amato G, Norell M, DeSalle R, Hayashi C. 2003. Combined support for wholesale taxic atavism in gavialine crocodylians. Syst. Biol. 52, 403–422. ( 10.1080/10635150350197037) [DOI] [PubMed] [Google Scholar]
- 2.Harshman J, Huddleston CJ, Bollback JP, Parsons TJ, Braun MJ. 2003. True and false gharials: a nuclear gene phylogeny of Crocodylia. Syst. Biol. 52, 386–402. ( 10.1080/10635150390197028) [DOI] [PubMed] [Google Scholar]
- 3.Martin JE, Buffetaut E, Naksri W, Lauprasert K, Claude J. 2012. Gavialis from the Pleistocene of Thailand and its relevance for drainage connections from India to Java. PLoS ONE 7, e44541 ( 10.1371/journal.pone.0044541) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jouve S., Bouyac B, Amaghzazc M, Meslouhd S. 2015. Maroccosuchus zennaroi (Crocodylia: Tomistominae) from the Eocene of Morocco: phylogenetic and palaeobiogeographical implications of the basalmost tomistomine. J. Syst. Palaeont. 13, 421–445. ( 10.1080/14772019.2014.913078) [DOI] [Google Scholar]
- 5.Brochu CA. 1997. Morphology, fossils, divergence timing, and the phylogenetic relationships of Gavialis. Syst. Biol. 46, 479–522. ( 10.1093/sysbio/46.3.479) [DOI] [PubMed] [Google Scholar]
- 6.Brochu CA. 2007. Morphology, relationships, and biogeographical significance of an extinct horned crocodile (Crocodylia, Crocodylidae) from the Quaternary of Madagascar. Zool. J. Linnean Soc. 150, 835–863. ( 10.1111/j.1096-3642.2007.00315.x) [DOI] [Google Scholar]
- 7.Brochu CA. 2004. A new Late Cretaceous gavialoid crocodylian from eastern North America and the phylogenetic relationships of thoracosaurs. J. Vert. Paleo. 24, 610–633. ( 10.1671/0272-4634(2004)024%5B0610:ANLCGC%5D2.0.CO;2) [DOI] [Google Scholar]
- 8.Lee MSY, Yates AM. 2018. Data from: Tip dating and homoplasy: reconciling the shallow molecular divergences of modern gharials with their long fossil record Dryad Digital Repository. ( 10.5061/dryad.3d23687) [DOI] [PMC free article] [PubMed]
- 9.Müller J, Reisz RR. 2005. Four well-constrained calibration points from the vertebrate fossil record for molecular clock estimates. Bioessays 27, 1069–1075. ( 10.1002/bies.20286) [DOI] [PubMed] [Google Scholar]
- 10.Oaks JR. 2011. A time-calibrated species tree of Crocodylia reveals a recent radiation of the true crocodiles. Evolution 65, 3285–3297. ( 10.1111/j.1558-5646.2011.01373.x) [DOI] [PubMed] [Google Scholar]
- 11.Green RE, et al. 2014. Three crocodilian genomes reveal ancestral patterns of evolution among archosaurs. Science 346, 1254449 ( 10.1126/science.1254449) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gold ME, Brochu CA, Norell MA. 2014. An expanded combined evidence approach to the Gavialis problem using geometric morphometric data from crocodylian braincases and eustachian systems. PLoS ONE 9, e105793 ( 10.1371/journal.pone.0105793) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiari Y, Cahais V, Galtier N, Delsuc F. 2012. Phylogenomic analyses support the position of turtles as the sister group of birds and crocodiles (Archosauria). BMC Biol. 10, 65 ( 10.1186/1741-7007-10-65) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benton MJ, Clark JM. 1988. Archosaur phylogeny and the relationships of the Crocodylia. In The phylogeny and classification of the tetrapods, vol. 1 (ed. Benton MJ.), pp. 295–338. Oxford, UK: Clarendon Press. [Google Scholar]
- 15.Swofford DL. 2003. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4. Sunderland, MA: Sinauer Associates. [Google Scholar]
- 16.Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. ( 10.1093/sysbio/sys029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973. ( 10.1093/molbev/mss075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ronquist F, Klopfstein S, Vilhelmsen L, Schulmeister S, Murray L, Rasnitsyn AP. 2012. A total-evidence approach to dating with fossils, applied to the early radiation of the Hymenoptera. Syst. Biol. 61, 973–999. ( 10.1093/sysbio/sys058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gavryushkina A, Heath TA, Ksepka DT, Stadler T, Welch D, Drummond AJ. 2017. Bayesian total-evidence dating reveals the recent crown radiation of penguins. Syst. Biol. 66, 57–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lanfear R, Calcott B, Ho SYW, Guindon S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29, 1695–1701. ( 10.1093/molbev/mss020) [DOI] [PubMed] [Google Scholar]
- 21.Rambaut A, Suchard MA, Xie D, Drummond AJ. 2014. Tracer v1.6. See http://beast.bio.ed.ac.uk/Tracer .
- 22.Nylander JAA, Wilgenbusch JC, Warren DL, Swofford DL. 2008. AWTY (are we there yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics 24, 581–583. ( 10.1093/bioinformatics/btm388) [DOI] [PubMed] [Google Scholar]
- 23.Brochu C. 2013. Phylogenetic relationships of Palaeogene ziphidont eusuchians and the status of Pristichampsus Gervais 1853. Earth Env. Trans. Roy. Soc. Edin. 103, 521–550. [Google Scholar]
- 24.Stein M, Hand SJ, Archer M. 2016. A new crocodile displaying extreme constriction of the mandible, from the late Oligocene of Riversleigh, Australia. J. Vert. Paleont. 36, e1179041 ( 10.1080/02724634.2016.1179041) [DOI] [Google Scholar]
- 25.Holliday CM, Witmer LM. 2009. The epipterygoid of crocodyliforms and its significance for the evolution of the orbitotemporal region of the eusuchians. J. Vert. Paleont. 29, 715–733. ( 10.1671/039.029.0330) [DOI] [Google Scholar]
- 26.Turner AH, Pritchard AC, Matzke NJ. 2017. Empirical and Bayesian approaches to fossil-only divergence times: a study across three reptile clades. PLoS ONE 12, e0169885 ( 10.1371/journal.pone.0169885) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee MSY, Palci A. 2015. Morphological phylogenetics in the genomic age. Curr. Biol. 25, R922-R929. ( 10.1016/j.cub.2015.07.009) [DOI] [PubMed] [Google Scholar]
- 28.O'Reilly JE, Donoghue PCJ. 2016. Tips and nodes are complementary not competing approaches to the calibration of molecular clocks. Biol. Lett. 12, 20150975 ( 10.1098/rsbl.2015.0975) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Norell MA, Novacek MJ. 1992. The fossil record and evolution: comparing cladistics and paleontologic evidence for vertebrate history. Science 255, 1690–1693. ( 10.1126/science.255.5052.1690) [DOI] [PubMed] [Google Scholar]
- 30.Benton MJ, Hitchin R, Willis MA. 1999. Assessing congruence between cladistics and stratigraphic data. Syst. Biol. 48, 581–596. ( 10.1080/106351599260157) [DOI] [PubMed] [Google Scholar]
- 31.King B, Qiao T, Lee MSY, Zhu M, Long JA. 2017. Bayesian morphological clock methods resurrect placoderm monophyly and reveal rapid early evolution in jawed vertebrates. Syst. Biol. 66, 499–516. ( 10.1093/sysbio/syw107) [DOI] [PubMed] [Google Scholar]
- 32.Fischer DC. 2008. Stratocladistics: integrating temporal data and character data in phylogenetic inference. Annu. Rev. Ecol. Evol. Syst. 39, 365–385. ( 10.1146/annurev.ecolsys.38.091206.095752) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Lee MSY, Yates AM. 2018. Data from: Tip dating and homoplasy: reconciling the shallow molecular divergences of modern gharials with their long fossil record Dryad Digital Repository. ( 10.5061/dryad.3d23687) [DOI] [PMC free article] [PubMed]
Data Availability Statement
Full details of the methods and results, along with datasets and analysis executables, have been deposited in the Dryad Digital Repository (http://dx.doi.org/10.5061/dryad.3d23687) [8].