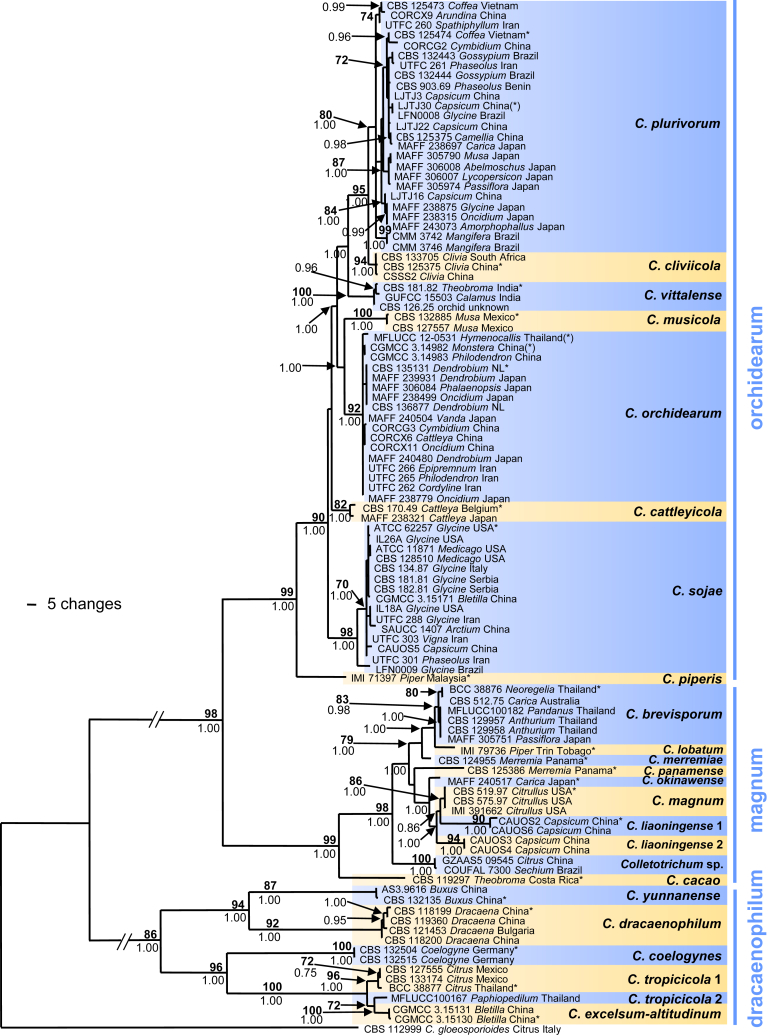
Fig. 1.
The first of 1 000 equally most parsimonious trees obtained from a heuristic search of the combined ITS, GAPDH, CHS-1, HIS3, ACT and TUB2 sequence alignment of the Colletotrichum dracaenophilum, C. magnum and C. orchidearum species complexes. Bootstrap support values above 70 % (bold) and Bayesian posterior probability values above 0.90 are shown at the nodes. Colletotrichum gloeosporioides strain CBS 112999 is used as outgroup. Numbers of ex-holotype and ex-epitype isolates are emphasised with an asterisk. Strain numbers are followed by substrate (host genus) and country of origin, NL = Netherlands, Trin Tobago = Trinidad and Tobago. Species complexes are indicated by blue lines. Branches that are crossed by diagonal lines are shortened by 50 %.