Figure 1. Identification of MOSPD2, a new FFAT motif‐binding protein.
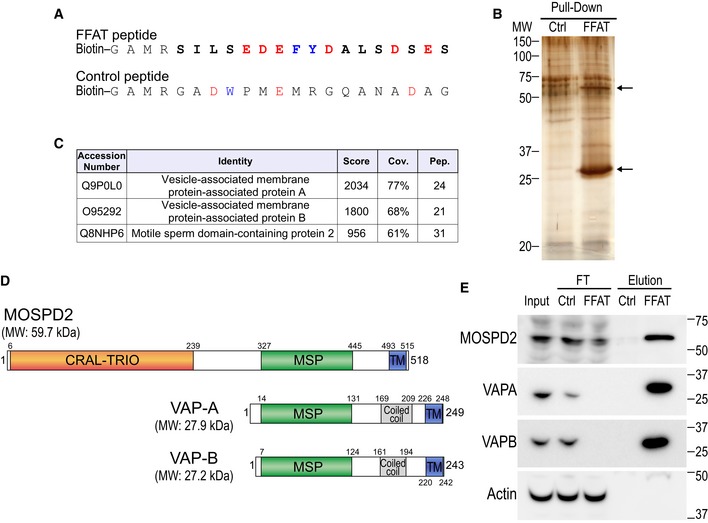
- Sequence of the two peptides used for the pull‐down assay. The peptides are composed of an amino‐terminal biotin, a linker sequence and the FFAT sequence of ORP1 (FFAT peptide) or a random sequence (control peptide). The FFAT sequence of ORP1 is in bold and corresponds to residues 469–483 (Accession Number Q9BXW6‐1). Aromatic and acidic residues are in blue and red, respectively.
- Silver nitrate staining of proteins pulled down using the control peptide (left) or the FFAT peptide (right), after SDS–PAGE. The two major differential bands are highlighted by arrows.
- Tandem mass spectrometry result table showing the three top‐scored proteins identified in the FFAT peptide‐bound fraction. Score: protein score based on the sum of the ion scores of all peptides identified; Cov.: percentage of the protein sequence covered by identified peptides; Pep.: number of unique peptide sequences identified.
- Schematic representation of MOSPD2, VAP‐A, and VAP‐B. Numbers correspond to the predicted positions of the beginning and the end of each domain. Calculated molecular weights (MW) of the proteins are shown.
- Western blot analysis of proteins pulled down using the control (Ctrl) peptide (left), or the FFAT (FFAT) peptide (right). The input, FT (flow through), and elution fractions correspond to HeLa cell total protein extract, unbound proteins, and bound proteins, respectively. Representative illustration of at least two independent experiments.
Source data are available online for this figure.
