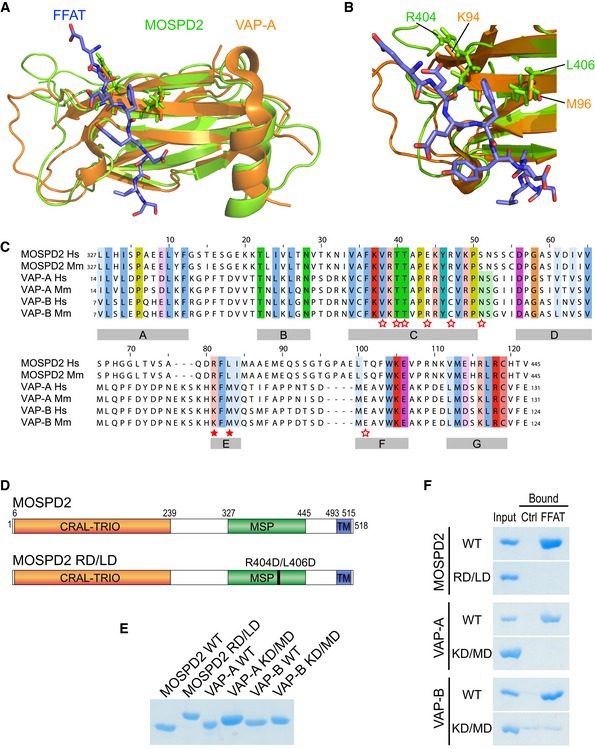
Figure 2. The MSP domain of MOSPD2 binds the FFAT motif.
-
A, B(A) Structural alignment of the MSP domains of MOSPD2 (PDB ID: 1WIC; green) and VAP‐A in complex with the FFAT motif of ORP1 (PDB ID: 1Z9O; orange). The FFAT peptide is in blue. (B) FFAT‐MSP binding interface: the critical residues K94, M96 in VAP‐A, and R404, L406 in MOSPD2 are shown as sticks.
-
CSequence alignment of the MSP domains of MOSPD2, VAP‐A, and VAP‐B from human (Hs) and mouse (Mm). Numbers refer to amino acid positions. Red stars indicate the position of residues interacting with the FFAT motif in VAP‐A 27. Filled stars indicate the position of the two critical residues in VAP proteins (K94 and M96 in VAP‐A; K87 and M89 in VAP‐B) for FFAT interaction. Subregions with high homology between the three proteins are indicated with gray boxes (A–G).
-
DSchematic representation of MOSPD2 showing the position of the RD/LD mutation of the MSP domain.
-
ECoomassie blue staining of recombinant wild‐type and mutant MSP domains of MOSPD2, VAP‐A, and VAP‐B on SDS–PAGE. Note that the mutant MSP domains of MOSPD2, VAP‐A, and VAP‐B displayed a slowed migration, likely resulting from the negative charges introduced by the mutations.
-
FCoomassie blue staining of proteins pulled down with the control or the FFAT peptide. Representative illustration of at least two independent experiments.
