Figure EV3. Multiple sequence alignments used in prediction of active site residues of Cas1 and DnaQ domains.
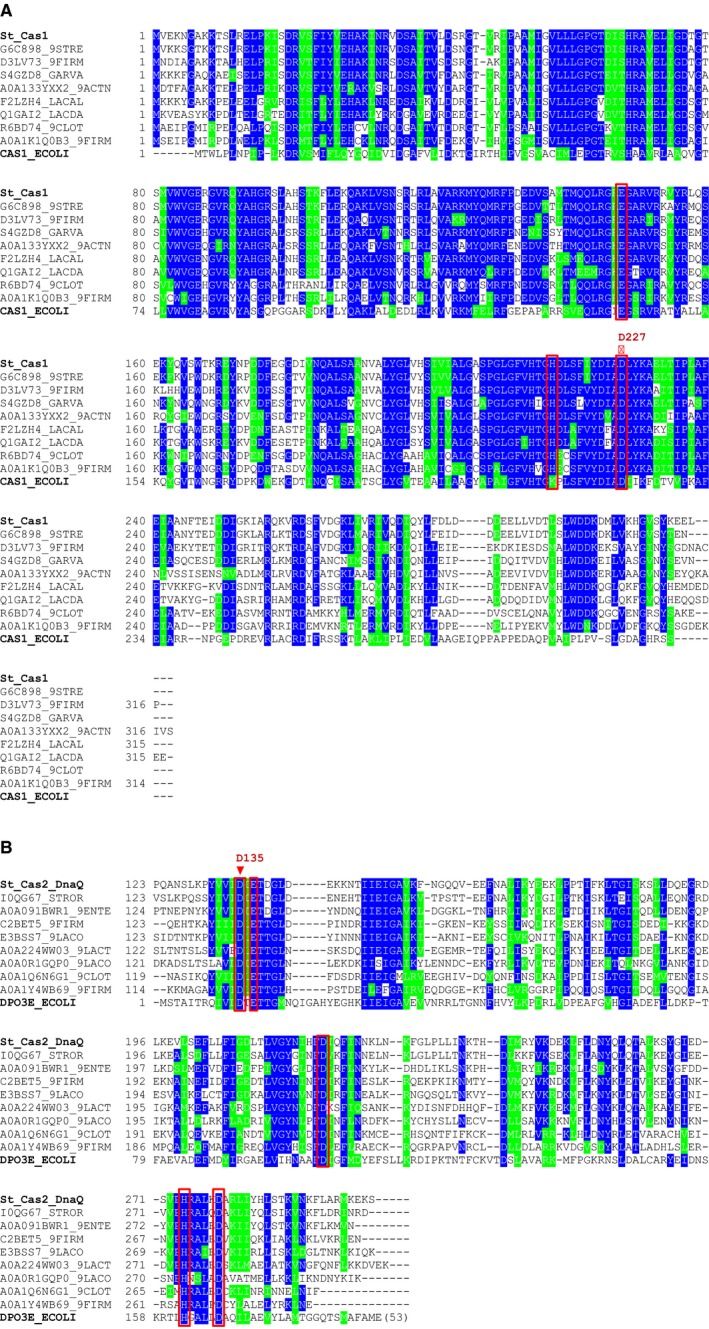
- Alignment of Streptococcus thermophilus Cas1 (St_Cas1) and its homologs including well‐characterized Escherichia coli Cas1 (CAS1_ECOLI). Positions corresponding to the active site residues of E. coli Cas1 are marked with red rectangles. The point mutation in S. thermophilus Cas1 is indicated with the red triangle. St_Cas1 homologs are represented by their Uniprot entry names.
- Alignment of S. thermophilus Cas2‐DnaQ (St_Cas2_DnaQ) and its homologs with E. coli PolIII ε‐subunit (DPO3E_ECOLI). Positions corresponding to the active site residues of ε‐subunit are marked with red rectangles. The amino acid that was mutated in S. thermophilus Cas2‐DnaQ is labeled with the red triangle. Sequences, except for St_Cas2_DnaQ, are represented by their Uniprot entry names.
