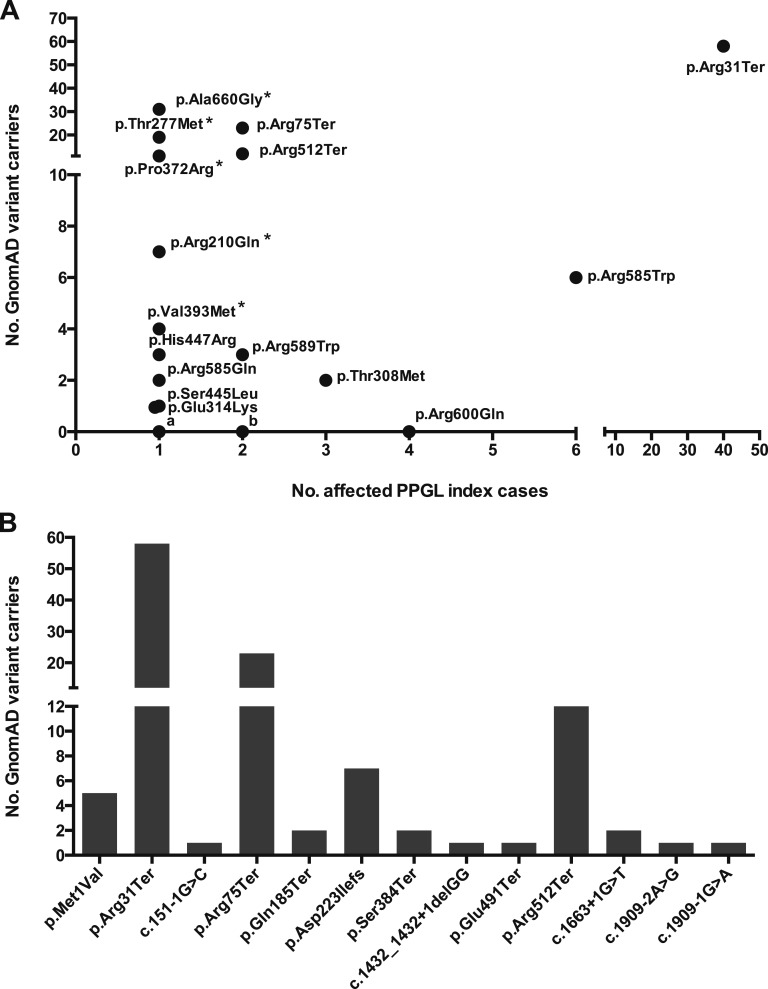
Figure 1.
Frequency of PPGL-associated SDHA variants in the GnomAD cohort. (A) Of the 39 individual SDHA variants reported in the literature in association with PPGL index cases, 15 (~40%) were observed in the GnomAD database. Strikingly, the p.Arg31Ter variant was observed at the highest frequency in both the PPGL and GnomAD cohorts. Several additional SDHA variants were observed recurrently in both disease and control cohorts (e.g., Arg585Trp, pThr308Met, p.Arg75Ter, p.Arg512Ter, p.Arg589Trp). In total, 183 individuals in the GnomAD database harbored one of the literature-associated PPGL-associated SDHA variants, representing ~1 in 750 of the GnomAD population. Excluding SDHA alleles reported as “variants of uncertain significance” in their original report (marked with an “*”), 111 GnomAD individuals harbored likely causative SDHA variants (i.e., ~1 in 1250). In contrast, 24 of the PPGL-associated SDHA variants were not observed in the GnomAD database. Of these, 19 variants were observed in single PPGL index cases (denoted “a”), 4 variants were observed twice (denoted “b”), and the p.Arg600Gln variant was observed in 4 unrelated PPGL cases. (B) All pathogenic or likely pathogenic LOF SDHA variants reported in the ClinVar database in association with PPGL/Hereditary Cancer Predisposition were identified and their frequency in the GnomAD cohort evaluated. In total, 33 putative LOF alleles were identified, of which 13 were observed in the GnomAD database (shown above). Overall, 116 individuals in the GnomAD cohort harbored one of the LOF SDHA alleles, equating to ~1 in 1200 of the cohort. All PPGL-associated variants were observed in the heterozygous state and are described relative to the canonical transcript ENST00000264932.