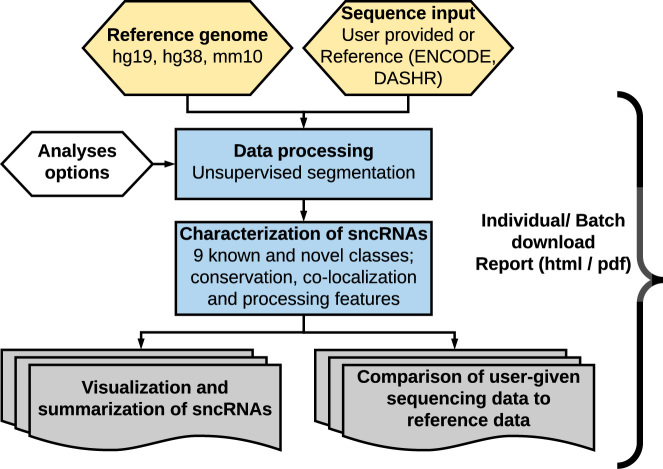
Figure 1.
The SPAR workflow. Given a reference genome and input small RNA-seq dataset (custom or reference data), SPAR processes the small RNA-seq dataset and identifies sncRNA loci using unsupervised segmentation. sncRNA loci are grouped into the major small RNA classes or the novel unannotated category (total of 10 classes) and annotated with various genomic features. The SPAR results are summarized in interactive tables at the per locus and genome-wide level and are compared with reference ENCODE and DASHR tissues and cell lines. All results are available for download in a variety of formats.