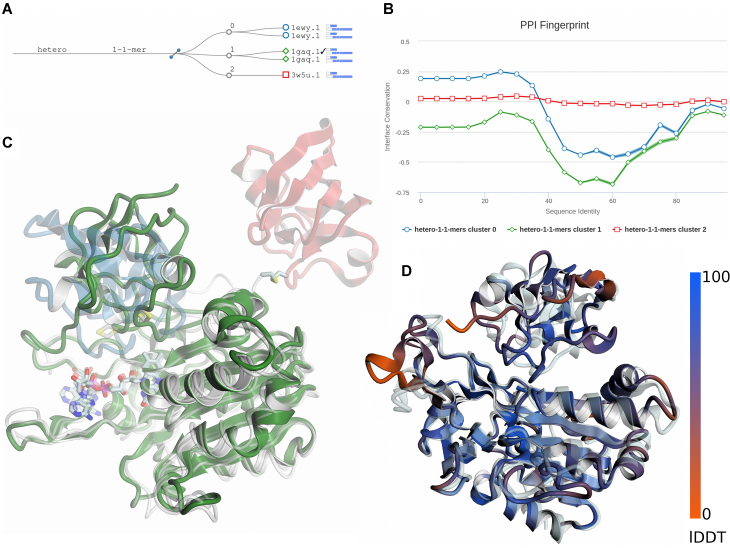
Figure 2.
Modelling example of the Ferrodoxin/Ferredoxin-NADP(+) Reductase hetero dimeric complex. (A) Decision tree of templates clustered according to their quaternary structure features: oligomeric state, stoichiometry, topology and interface similarity. Three different clusters are formed based on interface similarity between templates. (B) PPI fingerprint analysis of available template structures. The ratio between interface and surface residue entropy (interface conservation, y-axis) is reported as a function of evolutionary distance (sequence identity, x-axis). Templates corresponding to SMTL ID: 1ewy.1 (in blue) and SMTL ID: 1gaq.1 (in green) show the typical conservation pattern observed for biologically relevant interfaces, with stronger conservation signal in the sequence identity range between 40 and 60%. Considering also remote homologs (below 40% sequence identity), only the interface in template SMTL ID: 1gaq.1 is deemed as conserved (interface/surface conservation ratio below zero). Template corresponding to SMTL ID: 3w5u.1 (in red) displays an interface/surface conservation ratio close to zero, as observed in crystal contacts/artefacts. (C) Structure superposition of available templates. Each template is coloured according to same colouring scheme of Figure 2A and B. Templates corresponding to SMTL ID: 1ewy.1 (in blue) and 1gaq.1 (in green) show similar arrangement of FNR and Fd in the complex. Template SMTL ID: 3w5u.1 (red) shows a different localization of the Fd moiety. Cross-linked cysteines are shown in sticks. (D) Structure superposition between model and native structure of the root FNR:Fd complex. The model is coloured according to its local quality using a colour gradient from blue (high quality) to red (low quality) as measured by all-atom IDDT score. The native structure of the complex is shown in light gray.