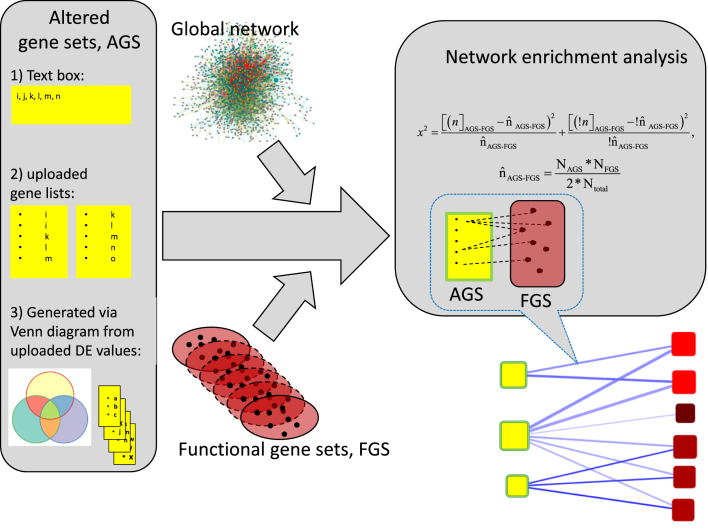
Figure 1.
Data flow of network enrichment analysis on EviNet.org. Three components shall be specified for an analysis on the server: (1) AGS, experimentally derived gene/protein lists in one of the three shown formats; (2) FGS, functional gene sets (typically pathways with well characterized biological function) selected from the FGS collection menu and (3) global network (also selected from the menu) where individual genes of AGS and FGS are supposed to be connected via edges (presence of all genes is not required, however AGS, FGS, and network must share the same name space). The program will identify available AGS-FGS edges, perform network enrichment analysis, produce graphical and tabular output, and store the results in Archive for future investigation. See details in ‘Data analysis and required input’.