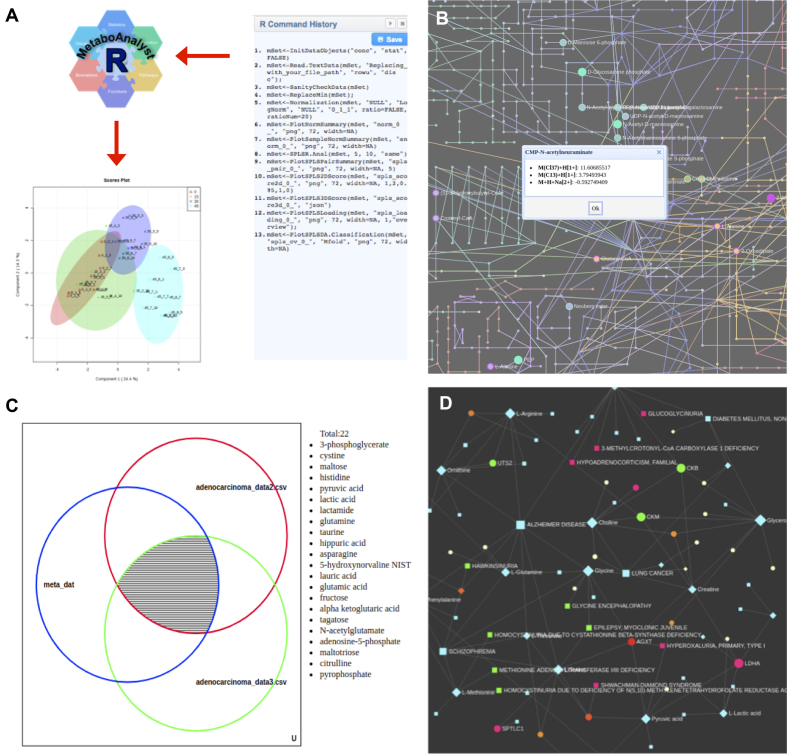
Figure 2.
Summary of new features introduced in MetaboAnalyst 4.0. (A) An illustration showing the R command history panel and the companion MetaboAnalystR package will allow users to easily reproduce their analyses. In this case, the R command history captures all R commands leading to the generation the PLS-DA 2D score plot, which can then be reproduced in MetaboAnalystR using identical R commands (except the file path parameter for user input). (B) A zoomed-in view of the KEGG metabolic network showing the potential metabolite hits predicted from the mummichog algorithm. Clicking on a highlighted node will display all possible matched adduct forms of the corresponding compound. (C) An interactive Venn diagram showing the results from a biomarker meta-analysis. Clicking on an area will show the corresponding hits. (D) An example of the metabolite-gene-disease interaction network created based on user input.