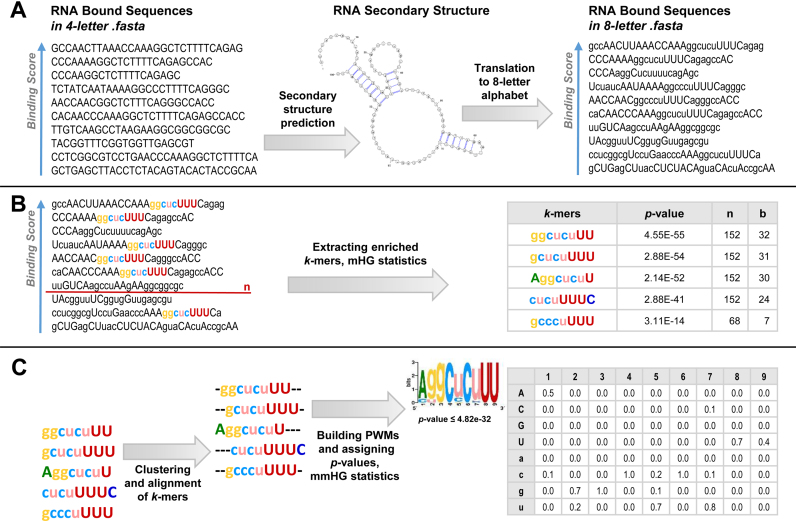
Figure 1.
A visualized summary of SMARTIV methodology. (A) The input for SMARTIV is a list of sequences ranked in a descending order according to the sequence binding scores. As a first step, we employ secondary structure predictions, defining each nucleotide in the sequence as either paired or unpaired and integrate the sequence and structural information to a new eight-letter alphabet (A,G,C,U for unpaired nucleotides and a,g,c,u for paired nucleotides). (B) We extract k-mers that are significantly enriched at the top of the ranked list compared to the bottom of the list, using the mHG statistics. (C) We cluster and align the k-mers. Consequently, we build a Position Weight Matrix (PWM) for each cluster, assigning it a P-value based on its correspondence to the original ranking of the sequences, based on the experimental binding scores, using the mmHG statistics.