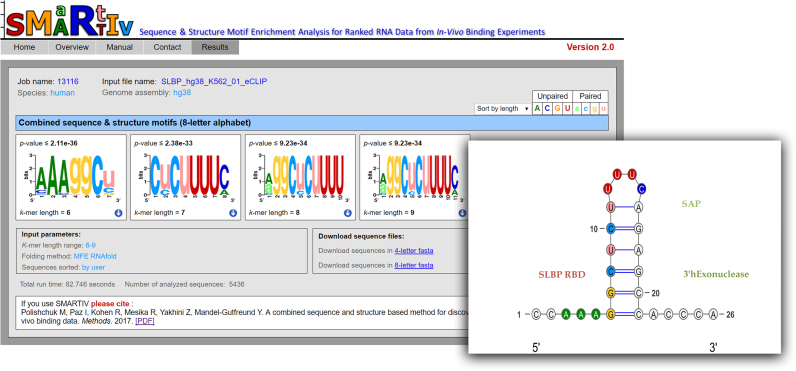
Figure 2.
SMARTIV results for extracting the combined sequence and structure motifs for SLBP. Rank list sequences from eCLIP experiment conducted for human SLBP in K562 cell were provided an input to SMARTIV webserver. Parameters were set to k-mer range: 6–9, and folding method: MFE RNAfold. Shown are the four most significant motifs in an eight-letter alphabet. On the right is a cartoon representing the secondary structure of the known SLBP binding motifs, which was solved by X-ray crystallography in complex with the SLBP protein. As shown, all four most significant motifs predicted by SMARTIV fit exactly to the known stem–loop binding site of SLBP on the histone mRNA, at both the sequence and structural level. For illustration, SMARTIV motif is mapped to the known stem–loop structure, using SMARTIV standard color-coding.