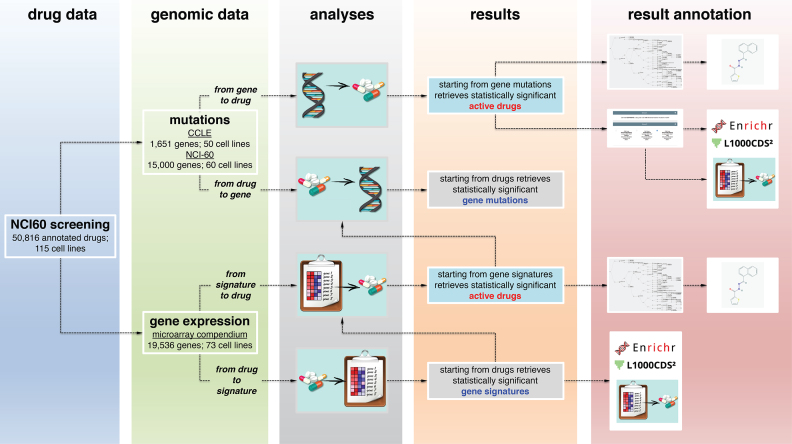
Figure 1.
Overview of GDA data analysis workflow. GDA is based on the pharmacological data obtained from the NCI-60 screening for a total of 50,816 compounds on 115 cancer cell lines and on the genomic and transcriptional profiles of the CCLE and NCI-60 studies. GDA can be interrogated through four main modules to identify drugs active in cancer cell lines bearing specific gene mutations (from gene to drug); gene mutations characterizing cancer cell lines that are responsive to a selected compound (from drug to gene); drugs active in cancer cell lines bearing the activation of a specific gene signature (from signature to drug); and up- and down-regulated genes in cancer cell lines that respond to a specific compound (from drug to signature). Results from the analyses can be fed into additional GDA modules (as the drug clustering, the Maximum Common Structure, and the differential gene expression analyses) or sent to external web services (as Enrichr, L1000CDS2 and PubChem) for functional annotation and comparison.