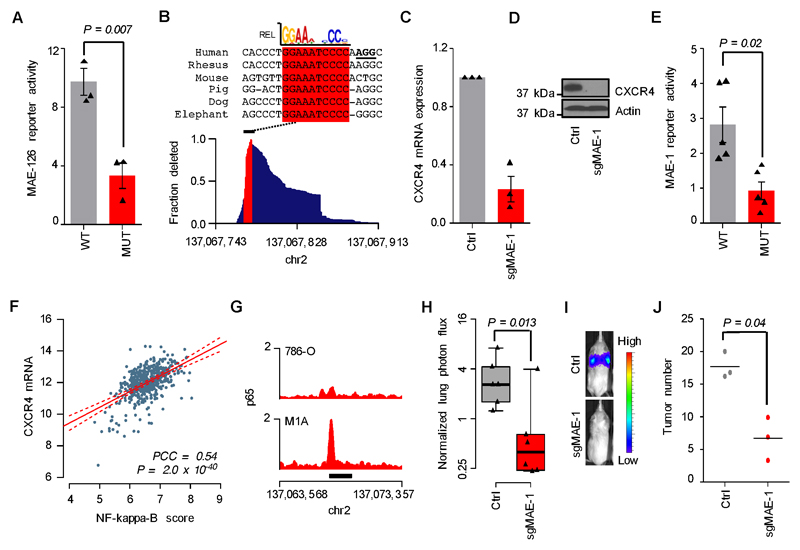
Figure 6. Mechanisms of MAE-1/MAE-126 activation in metastatic ccRCC.
A, Transient reporter assay showing the effects of mutated HIF motif on MAE-126 enhancer activity in M1A cells. Mean of three experiments. Error bars S.E.M. Two-sided Student’s t-test. B, Bottom, genomic MAE-1 deletion profile in M1A cells targeted by sgMAE-1. Top, mammalian sequence alignment of the most commonly deleted region, a conserved RELA/p65 motif highlighted in red. sgMAE-1 PAM sequence underlined in black. C, CXCR4 mRNA expression in M1A cells as measured by qRT-PCR. Mean of three experiments. Error bars S.E.M. D, CXCR4 protein expression in M1A cells as measured by immunoblotting. A representative experiment of three is shown. E, Transient reporter assay showing the effects of mutated RELA/p65 motif on MAE-1 enhancer activity in M1A cells. Mean of five experiments. Error bars S.E.M. Two-sided Student’s t-test. F, Correlation of CXCR4 mRNA expression with NF-kappaB activity in the TCGA ccRCC data set. N=506; PCC, Pearson’s correlation coefficient. G, p65 ChIP-seq data in the MAE-1 locus. Black bar indicates MAE-1. H, Effects of CRISPR/Cas9-mediated targeting of MAE-1 on lung metastatic fitness of M1A cells. Normalized lung photon flux in mice after 9 weeks of tail vein inoculation of 300,000 cells. N=6 for both groups. One-tailed Wilcoxon rank-sum test. Whiskers extend to data extremes. I, Representative bioluminescence images from the experiment shown in H. J, Histological quantification of lung metastatic foci from representative lungs of the experiment shown in H. N=3 for both groups. Horizontal bars represent sample mean. P-values for log-transformed data calculated by one-sided Student’s t-test.